当前位置:
X-MOL 学术
›
Environ. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
How good are global DNA‐based environmental surveys for detecting all protist diversity? Arcellinida as an example of biased representation
Environmental Microbiology ( IF 5.1 ) Pub Date : 2024-03-21 , DOI: 10.1111/1462-2920.16606 Fernando Useros 1 , Iván García‐Cunchillos 2 , Nicolas Henry 3, 4 , Cédric Berney 4 , Enrique Lara 1
Environmental Microbiology ( IF 5.1 ) Pub Date : 2024-03-21 , DOI: 10.1111/1462-2920.16606 Fernando Useros 1 , Iván García‐Cunchillos 2 , Nicolas Henry 3, 4 , Cédric Berney 4 , Enrique Lara 1
Affiliation
|
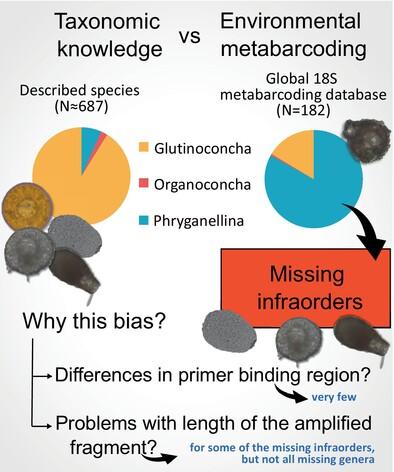
Metabarcoding approaches targeting microeukaryotes have deeply changed our vision of protist environmental diversity. The public repository EukBank consists of 18S v4 metabarcodes from 12,672 samples worldwide. To estimate how far this database provides a reasonable overview of all eukaryotic diversity, we used Arcellinida (lobose testate amoebae) as a case study. We hypothesised that (1) this approach would allow the discovery of unexpected diversity, but also that (2) some groups would be underrepresented because of primer/sequencing biases. Most of the Arcellinida sequences appeared in freshwater and soil, but their abundance and diversity appeared underrepresented. Moreover, 84% of ASVs belonged to the suborder Phryganellina, a supposedly species‐poor clade, whereas the best‐documented suborder (Glutinoconcha, 600 described species) was only marginally represented. We explored some possible causes of these biases. Mismatches in the primer‐binding site seem to play a minor role. Excessive length of the target region could explain some of these biases, but not all. There must be some other unknown factors involved. Altogether, while metabarcoding based on ribosomal genes remains a good first approach to document microbial eukaryotic clades, alternative approaches based on other genes or sequencing techniques must be considered for an unbiased picture of the diversity of some groups.
中文翻译:

全球基于 DNA 的环境调查对于检测所有原生生物多样性有多好? Arcellinida 作为有偏见的代表性的一个例子
针对微真核生物的元条形码方法深刻改变了我们对原生生物环境多样性的看法。公共存储库 EukBank 由来自全球 12,672 个样本的 18S v4 元条形码组成。为了估计该数据库在多大程度上提供了所有真核生物多样性的合理概述,我们使用Arcellinida(叶状睾丸变形虫)作为案例研究。我们假设(1)这种方法可以发现意想不到的多样性,但(2)由于引物/测序偏差,某些群体的代表性不足。大多数Arcellinida序列出现在淡水和土壤中,但它们的丰度和多样性似乎代表性不足。此外,84% 的 ASV 属于 Phryganellina 亚目,这是一个被认为是物种匮乏的进化枝,而记录最齐全的亚目(Glutinoconcha,已描述的 600 个物种)仅具有少量代表性。我们探讨了这些偏见的一些可能原因。引物结合位点的错配似乎起着次要作用。目标区域过长可以解释其中一些偏差,但不是全部。一定还有其他一些未知的因素参与其中。总而言之,虽然基于核糖体基因的元条形码仍然是记录微生物真核进化枝的良好首选方法,但必须考虑基于其他基因或测序技术的替代方法,以公正地了解某些群体的多样性。
更新日期:2024-03-21
中文翻译:

全球基于 DNA 的环境调查对于检测所有原生生物多样性有多好? Arcellinida 作为有偏见的代表性的一个例子
针对微真核生物的元条形码方法深刻改变了我们对原生生物环境多样性的看法。公共存储库 EukBank 由来自全球 12,672 个样本的 18S v4 元条形码组成。为了估计该数据库在多大程度上提供了所有真核生物多样性的合理概述,我们使用Arcellinida(叶状睾丸变形虫)作为案例研究。我们假设(1)这种方法可以发现意想不到的多样性,但(2)由于引物/测序偏差,某些群体的代表性不足。大多数Arcellinida序列出现在淡水和土壤中,但它们的丰度和多样性似乎代表性不足。此外,84% 的 ASV 属于 Phryganellina 亚目,这是一个被认为是物种匮乏的进化枝,而记录最齐全的亚目(Glutinoconcha,已描述的 600 个物种)仅具有少量代表性。我们探讨了这些偏见的一些可能原因。引物结合位点的错配似乎起着次要作用。目标区域过长可以解释其中一些偏差,但不是全部。一定还有其他一些未知的因素参与其中。总而言之,虽然基于核糖体基因的元条形码仍然是记录微生物真核进化枝的良好首选方法,但必须考虑基于其他基因或测序技术的替代方法,以公正地了解某些群体的多样性。































 京公网安备 11010802027423号
京公网安备 11010802027423号