Effectiveness of a novel compound microbial agent for water quality and the gut microbiota of Micropterus salmoides
- 1Aquaculture Department, Zhejiang Institute of Freshwater Fisheries, Huzhou, Zhejiang, China
- 2Aquaculture Department, Agriculture and Rural Affairs Key Laboratory of Healthy Freshwater Aquaculture, Huzhou, Zhejiang, China
- 3Aquaculture Department, Key Laboratory of Aquatic Product Quality Improvement and Processing Technology, Huzhou, Zhejiang, China
- 4Institute of Marine Biology, Ocean Research Center, Zhejiang University, Zhoushan, Zhejiang, China
The cultivation of largemouth bass (Micropterus salmoides), a species of significant economic value in aquaculture, has experienced notable growth recently. However, the deterioration of water quality seriously affects the metabolic responses of M. salmoides. While compound microbial agent (CMA) is widely utilized for ecological rehabilitation and water filtration, its application in M. salmoides has not been reported. Here, based on physio-biochemical tests and 16S rRNA sequencing, we investigated the effects of CMA (yeast, Bacillus subtilis, and lactic acid bacteria) on the water quality within the recirculating aquaculture system, along with physiological indices and gut microbiota of M. salmoides. Compared to the control and single microbial agent (yeast), CMA treatment improved the water quality by improving the dissolved oxygen and delaying the increase of pH, total nitrogen, total phosphorus, ammonia nitrogen, and nitrite. The 16s rRNA gene sequencing revealed that the water treated with CMA exhibited elevated levels of chao1, Shannon, Pd, and a larger population of dominant bacterial. Besides, higher values of ACE, chao1, Shannon, and OTU level, and lower Simpson index were found in CMA treated M. salmoides samples, suggesting that CMA treatment enhanced the species richness and diversity of gut microbiota of M. salmoides. Furthermore, CMA treatment hindered the generation and proliferation of harmful bacteria, such as the Mycoplasma mobile 163K species and the Erysipelotrichaceae family, which was associated with enhanced antioxidant enzymatic activity and decreased MDA level in both the serum and liver. These findings shed light on the essential roles of CMA in M. salmoides culturing and introduce an innovative approach to enhance the aquatic environment.
1 Introduction
Various microorganisms such as viruses, bacteria, and fungi exhibit significant potential in combating pathogens, thus serving as viable alternatives for environmentally burdensome compounds as biological control agents (Zehra et al., 2021). In plant biology, the utilization of microbial agents holds promising prospects in effectively regulating the progression of crop diseases by engaging with pathogenic bacteria or plant hosts, thereby bolstering plant resistance against stress (Maciag et al., 2023). The utilization of compound microbial agent (CMA) in animal production serves as an alternative to antibiotics by addressing the deficiency of beneficial microorganisms in the animal intestine, triggering the activation of the intestine immune system, and alleviating oxidative reactions (Liang et al., 2020). Within the aquaculture production, the adoption of commercial microbial agents, including probiotics, microbial products, and microbial preparations, has been extensively employed in shrimp culturing to prevent and control diseases, demonstrating their capacity to expedite the substitution of plankton bacterial communities, and the transmission of gut microbiota (Liu et al., 2018). Additionally, compared with conventional physical techniques, like mechanical filtration, and chemical methods, such as ozone treatment, microbial agent treatment offers distinct advantages of cost-effectiveness and minimal environmental impact (Zhai et al., 2020).
Generally, concentrations of ammonia nitrogen and nitrite are relatively low in natural aquatic environments but are elevated in high-density aquaculture systems as a result of anthropogenic activities and ecological degradation (Yang et al., 2017). Within aquaculture systems, ammonia nitrogen permeates aquatic organisms via the biofilm, gills, and mucous membranes, resulting in adverse effects on the central nervous system, thereby impacting the feeding and growth processes of aquatic creatures, rendering them more prone to diseases (Edwards et al., 2024). Upon entering the bloodstream through the gill filament, nitrite leads to a decrease in the quantity of red blood cells and hemoglobin in aquatic organisms, resulting in oxygen-deficient mortality during metabolic activity (Dutra et al., 2017). Therefore, the ability of microbial agents to lower levels of ammonia nitrogen and nitrite in water underscores their importance in improving water quality in densely populated aquaculture systems (Ren et al., 2021).
In addition to water quality, the health of aquatic creatures can be enhanced through the application of microbial agents (Giri et al., 2020). These agents have been shown to increase survival rates and maintain overall health by preserving gut barrier integrity, balancing microbial ecosystem within the gut, boosting immune responses, regulating nutrient absorption, and effectively absorbing and degrading excessive antibiotics (Narayanan et al., 2023). It has been reported that assessing changes in gut microflora can serve as a valuable indicator of the health status of aquaculture organisms (El-Saadony et al., 2021). The gut microbiota inhabiting the gastrointestinal tract of the host engage in a symbiotic relationship, producing metabolites that play a crucial role in the health by influencing processes such as digestion, metabolism, and immune response (Ni and Tokuda, 2013). The species Micropterus salmoides is extensively cultivated worldwide for its nutritional value and rapid growth rate (Niimi and Beamish, 1974). M. salmoides have garnered favor among farmers and consumers due to their wide temperature adaptability, robust disease resistance, short aquaculture cycle, delectable meat, and ease of capture (Yin et al., 2022). Additionally, MsTRIM21 of M. salmoides has the capacity to markedly impede viral replication while amplifying immune and inflammatory reactions of the host, thereby assuming a pivotal function in innate immune response against iridovirus (Peng et al., 2023). However, the cultivation of M. salmoides is increasingly threatened by heavy metal pollution and eutrophication in water bodies (Julian and Gu, 2015). Besides, although the beneficial effects of CMA have been demonstrated in plant disease resistance (Chen et al., 2022) and shrimp culturing (Liu et al., 2018), its application in M. salmoides culturing has not been reported.
To investigate the potential of microbial agents in enhancing water quality and the health of M. salmoides, M. salmoides were fed with commercial food containing microbial agents, and we determined the impact of microbial agents on pH, dissolved oxygen (DO), the accumulation of total nitrogen, total phosphorus, ammonia nitrogen, and nitrite, as well as changes in the structure and composition of the microbial community in the water samples. Besides, we measured changes in the composition of gut microbiota, the activity of antioxidant enzyme, and the growth indices of M. salmoides. Overall, this study sheds light into the application of microbial agents on M. salmoides aquaculture and provides a novel approach to improve water quality.
2 Materials and methods
2.1 Preparation of fish sample and microbial agents
Juvenile fry of M. salmoides were obtained from the freshwater aquaculture base of Zhejiang Institute of Freshwater Research (29.9°S, 122.4°E), Zhejiang Province, China. The samples were acclimated in the laboratory for a duration of 14 d and were fed with a commercial diet (Catalog No. 0719, purchased from Guangzhou Muchang Biotechnology Co., Ltd.) containing protein (42%) and fat (8%). A total of 540 individuals with consistent size (29.54 ± 2.07 g) were randomly allocated to nine aquariums (60 samples per aquarium) equipped with a recirculation system featuring a biological filter, and each aquarium held a capacity of 100 L with a flow rate of 2 L/min. To preserve the aquatic environment quality, a daily exchange of 10% of the culture water was conducted throughout the entire breeding phase. The water temperature was maintained at 25°C, with continuous oxygenation ensured that the dissolved oxygen levels were above 5.0 mg/L, while ensuring ammonia nitrogen levels remained below 0.02 mg/L and a pH of 7.0 ± 0.5. The feeding trial lasted for a duration of 8 weeks, during which M. salmoides samples were fed twice daily until reaching satiety. The amount of feed consumed and the number of sample mortalities in each aquarium were recorded at regular intervals.
To investigate the role of microbial agents in maintaining water quality and the health of M. salmoides samples, following the 14-day pre-culture period, we introduced a single microbial agent (yeast, Catalog No. BJJM-7001, purchased from Shandong Huamo Biotechnology Co., Ltd.) (SMA) and CMA (yeast, Bacillus subtilis, Catalog No. RY-JF001, purchased from Guangdong Ranyi Biotechnology Co., Ltd., and lactic acid bacteria, Catalog No. LA71, purchased from Shandong Huamo Biotechnology Co., Ltd., 1:1:1) to the diet (came from Zhoushan Marine Research Center of Zhejiang University) and provided it to M. salmoides samples (Zhou et al., 2020). Among them, yeast was selected as a point of general comparison in this study owing to its prevalent application in aquaculture (Øverland and Skrede, 2017). The samples in the control group were administered the diet without any microbial agents, and treatment groups were fed with SMA (4 g) and CMA (4 g) every 7 d (Guo et al., 2020). In each experiment, except for the microbial agents in the feed, M. salmoides samples from the control, SMA, and CMA groups were subjected to exactly the same conditions. Each trial consisted of three technical replicates and three biological replicates.
2.2 Determination of water quality
To assess the impact of microbial agents on improving the water quality of the aquarium, from the 1st day after the treatment of SMA and CMA, we randomly collected three parallel samples using a 50 mL syringe at a depth of 50 cm and mixed as the water samples (1 L). The HACH HQD Portable Meter (HQ40d, HACH) was used to measure the pH and DO levels (Sun et al., 2023). Additionally, the levels of ammonia nitrogen and nitrite in the aquaculture circulation system were determined using the standard colorimetric method (Valencia-Castañeda et al., 2019). The contents of total nitrogen (TN) and total phosphorus (TP) were assayed using the method (Zhang et al., 2023).
To clarify the correlation between microbial community structure and the additional microbial agents, we employed approach based on 16S ribosomal RNA (16S rRNA) gene sequencing using the Illumina MiSeq platform (Guangzhou Genedenovo Biotechnology Co., Ltd.). D0 represented the initial water sample before microbial agent treatment, D13-CK represented the water sample of the control group at 13 d, D13-S represented the water sample of SMA treatment group at 13 d, D13-C represented the water sample of CMA treatment group at 13 d, D25-CK represented the water sample of the control group at 25 d, D25-S represented the water sample of SMA treatment group at 25 d, D25-C represented the water sample of CMA treatment group at 25 d, D32-CK represented the water sample of the control group at 32 d, D32-S represented the water sample of SMA treatment group at 32 d, D32-C represented the water sample of CMA treatment group at 32 d, respectively. Each water sample was processed through filtration using a 0.22 μm microporous filter membrane within 4 h. Subsequently, the microbial genomic DNA was extracted using the bacterial DNA extraction kit (Catalog No. 9182, purchased from Beijing Takara Biotechnology Co., Ltd.), followed by the amplification of the target region of 16s utilizing specific primers (specific primer sequence, 338F: 5’-ACTCCTACGGGAGGCAGCAG-3’, 806R: 5’-GGACTACHVGGGTWTCTAAT-3’) with the barcode. Additionally, the PCR annealing temperature used for 16s rRNA was 52°C. The resulting PCR products were then analyzed through gel electrophoresis and recovery, with quantification performed using Qubit3.0. The amplified products were amalgamated in equal proportions, and the sequencing adaptors were attached to establish the sequencing library.
2.3 Growth conditions and antioxidant capacity of M. salmoides
To assess the growth conditions of M. salmoides under different treatments, following an 8-week treatment period, a total of nine M. salmoides samples from each treatment group were randomly selected from three parallel aquaria. These selected samples, three from each aquarium, were anesthetized with MS-222 at a concentration of 10 mg/L. Growth performance and feed utilization were determined using a previous method (Guo et al., 2020). Additionally, the formula used for determination of survival rate (SR), final length (FL), final weight (FW), relative gain rate of body weight (RGRBW), hepatosomatic index (HIS), visceral index (VSI), and fatness ratio (FR) was referred to the previous literature (Che et al., 2023).
The blood was meticulously obtained from the tail vein using a disposable syringe. The blood was then carefully transferred into a centrifuge tube, stood for 2 h at 4°C. Following this, the serum was separated from the blood through centrifugation at 4000 rpm for 10 min at 4°C. This serum was then utilized for determining antioxidant enzyme activity. The protein concentration of the homogenates was determined using the method (Bradford, 1976). The activities of superoxide dismutase (SOD), catalase (CAT), and GSH-peroxidase (GSH-Px) in M. salmoides serum was determined using the SOD Activity Assay Kit (Catalog No. BC5165), CAT Activity Assay Kit (Catalog No. BC0205), and GSH-Px Activity Assay Kit (Catalog No. BC1190), respectively. Malondialdehyde (MDA) content of M. salmoides serum was assayed using MDA Content Assay Kit (Catalog No. BC0025). Furthermore, all these kits were purchased from Beijing Solarbio Technology Co., Ltd.
Additionally, the experimental procedure was approved by the Experimental Animal Ethics Committee of Zhejiang Institute of Freshwater Fisheries, and all aquatic animal experiments strictly complied with the Experimental Animal Management Ordinance of Zhejiang Province, China.
2.4 16S rDNA sequencing of gut microbiota in M. salmoides
The intestines of anesthetized M. salmoides samples were expeditiously dissected and removed. Subsequently, the excised intestines were promptly frozen in liquid nitrogen and stored at -80°C for 16S rDNA sequencing of the gut microbiota.CK represented the gut microbial community of M. salmoides fed with the diet without microbial agents, S represented the gut microbial community of M. salmoides fed with single microbial agent, and C represented the gut microbial community of M. salmoides fed with CMA. To extract the total genomic DNA from the gut samples, MiniBEST Whole Blood Genomic DNA Extraction Kit (Catalog No. 9781, purchased from Beijing Takara Biotechnology Co., Ltd.) was employed. The extracted DNA was detected using 1% agarose gel electrophoresis. To establish a total of twelve libraries, PCR amplification was performed using specific primers (341F: 5’-ACTCCTACGGGAGGCAGCAGCAG-3’, 806R: 5’-GGACTACHVGGGTWTCTAAT-3’) with a annealing temperature of 52°C (Hugerth et al., 2014).
Agarose gel electrophoresis was employed to detect PCR products. Subsequently, the Illumina platform was used to sequence the sample, with the FLASH merge match terminal being employed for data reading. To obtain high-quality clean data, the raw data underwent qualitative filtering under specific conditions using the FQTRIM software (version. 0.9.4) (Heikema et al., 2020). Statistical analysis was conducted using QIIME2 software and R software (version. 3.5.2) (Chen et al., 2023). Vsearch9 (version. 2.3.4) software was utilized to verify and eliminate chimera sequences (Johnson et al., 2019). QIIME2 software was employed to assess the composition and abundance distribution of each sample at five classification levels, such as phyla, class, order, family, and genus. Additionally, alpha-diversity indices were calculated for each sample, including chao1, observed species, Shannon, Simpson index, and coverage rate (Dan et al., 2020).
2.5 Statistical analyses
Statistical analysis utilizing t-test, one-way or two-way ANOVA, and GraphPad software (version 8.2.1) was conducted to evaluate significant differences in biochemical indices among the control, SMA and CMA treatment groups of M. salmoides. In addition, the P value of ≤ 0.05 and ≤ 0.01 were classified as significant (*) and very significant (**), respectively.
3 Result
3.1 Improvement of water quality
To investigate the effects of microbial agents on the water quality of M. salmoides aquaculture system, we determined the pH and DO values over a period of 7 d following treatment with SMA and CMA. The results showed that during first 3 days, there was no significant difference between ddH2O, SMA, and CMA treatments. However, in the subsequent 4 d treatment, the pH values of M. salmoides aquaculture system under SMA and CMA treatments were significantly lower than that of ddH2O treatment group, with CMA treatment displaying the lowest pH (Figure 1A). In contrast to pH, the DO values in these groups exhibited an initial decline followed by an increase. Compared to the control group, the DO values in both the SMA and CMA treatment groups exhibited an increase, with the CMA treatment group demonstrating significantly higher DO levels at 6 d (1.02-fold increase) and 7 d (1.05-fold increase) (Figure 1B).
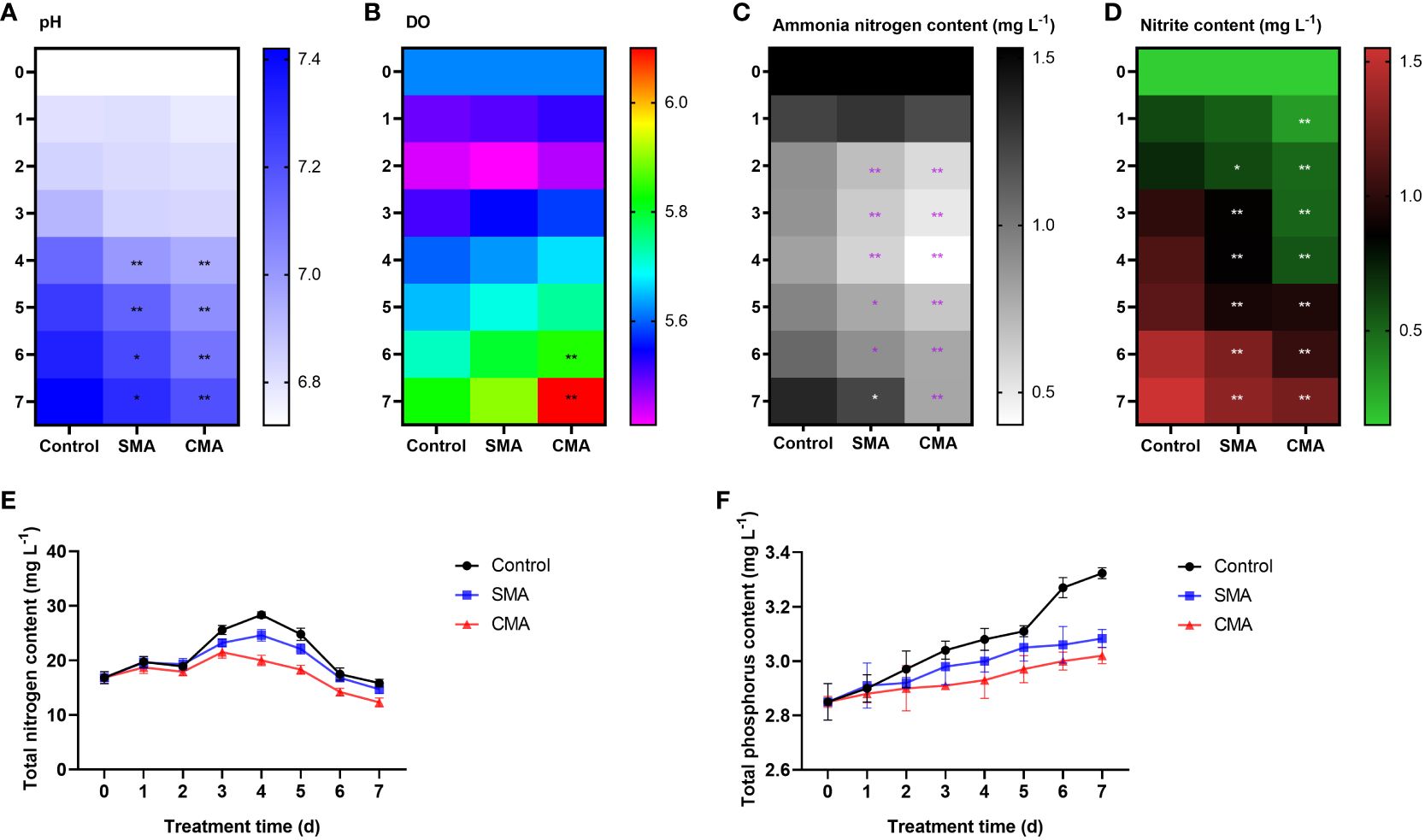
Figure 1 The effects of microbial agents on the physical-chemical properties of aquatic water in M. salmoides aquaculture system. (A) pH, (B) DO, (C) ammonia nitrogen content, (D) nitrite content, (E) TP content, and (F) TN content in the control, SMA, and CMA treated water samples. *P ≤ 0.05 and **P ≤ 0.01 were calculated by two-way ANOVA compared with the control.
To explore the impact of microbial agents on eutrophication in M. salmoides aquaculture system, we determined the contents of total nitrogen, total phosphorus, ammonia nitrogen, and nitrite in different treatment groups. The results showed that in comparison to the control group, the contents of ammonia nitrogen (Figure 1C), nitrite (Figure 1D), total phosphorus (Figure 1E), and total nitrogen (Figure 1F) were significantly lower in SMA and CMA groups. Noticeably, the water quality indicators of CMA treated group was better than that of SMA treated group, indicating that compared to SMA treatment, CMA treatment was more effective in maintaining water quality of M. salmoides aquaculture system.
3.2 Effect of microbial diversity in M. salmoides
The sequencing data revealed that a total of 4,617,867 high-quality reads were obtained from 36 water samples, with an average of 128,274 reads per sample. These reads demonstrated an average sequence length of 457 base pairs. Furthermore, the analysis of species annotation indicated the presence of 29 phyla, 67 classes, 192 orders, 320 families, 556 genera, and 861 species within the water samples. The evaluation of chao1 index, which measured the flora abundance, revealed that the control group exhibited a peak chao1 index at 25 d before a decline at 32 d. In contrast, the water sample treated with CMA displayed a higher chao1 index compared to both the control and SMA treated groups. Furthermore, the chao1 index of the CMA treated group at 32 d was significantly higher than that of the control group (Figure 2A). Meanwhile, it was observed that although there was no significant difference among these groups, the Shannon index (Figure 2B) and Pd index (Figure 2C) of the three groups showed a similar pattern to the chao1 index, suggesting that the microbial diversity in the CMA treatment group was higher than that of both the SMA treatment and the control group.
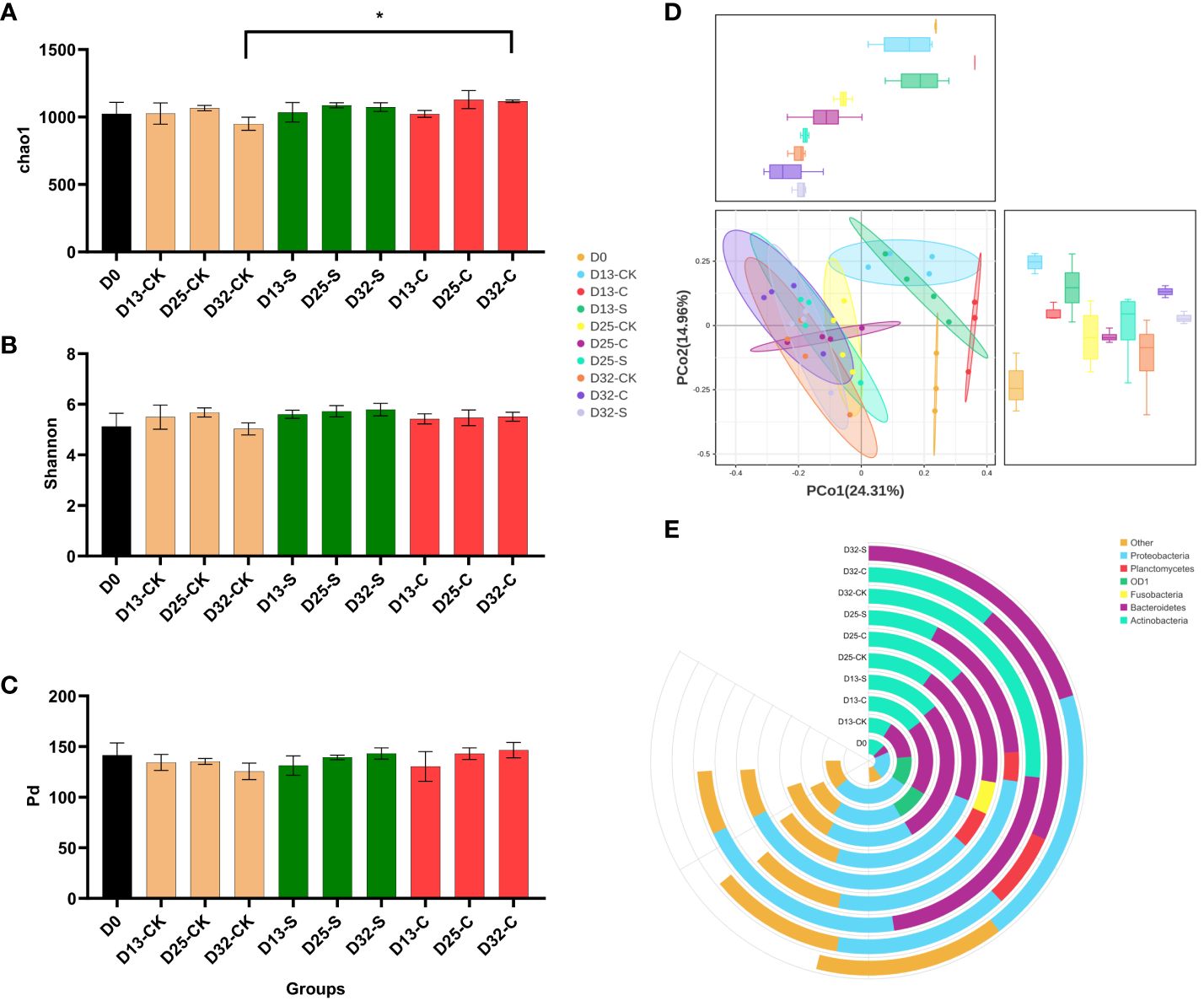
Figure 2 Comparison of diversity and bacterial composition of water samples based on 16S rRNA sequencing database. (A) chao1 index, (B) Shannon index, (C) Pd index, (D) PCoA of the microbial composition in the control, SMA, and CMA treated water samples. (D) Relative abundance of the top bacterial genera in the control, SMA, and CMA treated water samples. *P ≤ 0.05 was calculated by one-way ANOVA compared with the control.
Based on the 556 genera abundance, the principal coordinate analysis (PCoA) found that PC1 and PC2 accounted for 24.31% and 14.96%, respectively. The initial water group exhibited distinct separation from the other treatment groups, and the treatment groups at 13 d were distinct from those at 25 d and 32 d, suggesting that the alteration in the abundance of the microbial community was accompanied by the duration of the treatment period (Figure 2D). Moreover, the stacked graph of microbes in major phyla showed that Actinobacteria, Bacteroidetes, and Proteobacteria were enriched across all samples during the culture period. Notably, the gut flora of CMA treatment group exhibited higher diversity in terms of richness compared to the control group and SAM treatment group at 32 d (Figure 2E). Based on the 861 species abundance with a linear discriminant analysis (LDA) score threshold for 3.0, linear discriminant analysis effect size (LEFSe) analysis found that at 32 d, CMA treatment group showed a significant greater number of influential bacteria (Sun et al., 2023) than the control group (Liu et al., 2018) and SAM treatment group (Dutra et al., 2017) (Figure 3A). Among these treatment groups, the dominant bacterial species in the control group included Lacihabitans genus (Bacteroidetes phylum), Aurantimicrobium genus (Actinobacteria phylum), Hyalella azteca species (Bacteroidetes phylum), Sphingobacteriaceae family (Bacteroidetes phylum), Actinobacteria class (Actinobacteria phylum), and Microbacteriaceae family (Actinobacteria phylum), the dominant bacterial species in SMA group included Armatimonas genus (Armatimonadetes phylum) and Armatimonadales order (Armatimonadetes phylum), while the dominant bacterial species in CMA group were Microtrichales order (Actinobacteria phylum), Phycisphaerae class (Planctomycetes phylum), Arenimonas genus (Proteobacteria phylum), Phycisphaerales order (Planctomycetes phylum), Reyranellaceae family (Proteobacteria phylum), Gemmatales order (Planctomycetes phylum), Mycoplasma moatsii ATCC 27625 species (Tenericutes phylum), Steroidobacterales order (Proteobacteria phylum), and so on. Furthermore, the result of gut flora function prediction showed that the relative abundances of specific beneficial bacterial strains, such as Firmicutes, Bacteroidetes, and Actinobacteria, were higher in CMA treated M. salmoides samples (Figure 3B).
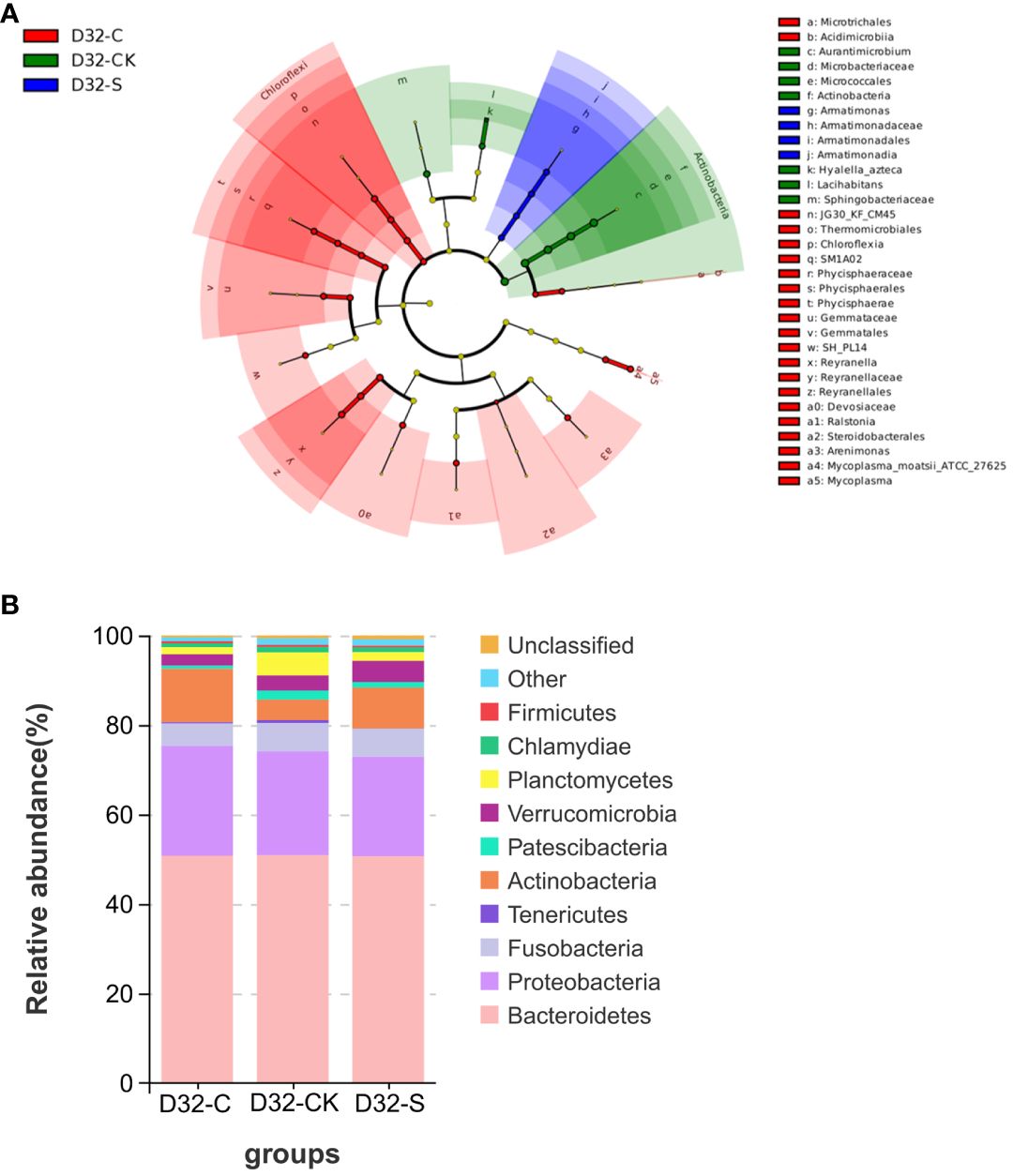
Figure 3 Differential bacterial genera identified among different treatment groups. (A) LEFSe analysis, and LDA score threshold was set as 3.0. The green group represented the control water sample at 32 d, the blue group represented SMA treated water sample at 32 d, and the red group represented CMA treated water sample at 32 d, respectively. (B) Relative abundance of the top 10 bacterial of M. salmoides under control, SMA, and CMA treatments at 32 d (at phylum level).
3.3 Improvement in growth and antioxidant enzymatic activity
To investigate the impact of water quality and microbial agents on the survival and favorable growth of M. salmoides, the survival rate and growth performance of M. salmoides were evaluated over a period from feeding initiation day to 41-day. As presented in Table 1, no significant differences were observed in the relative growth rate of body length (RGRBL), feed conversion ratio (FCR) among M. salmoides fed with the control feed, SAM and CMA. Meanwhile, compared to the control, the two groups fed with SMA and CMA differed significantly in regard to most growth indicators, including survival rate (SR), final length (FL), final weight (FW), relative gain rate of body weight (RGRBW), hepatosomatic index (HIS), visceral index (VSI), and fatness ratio (FR).
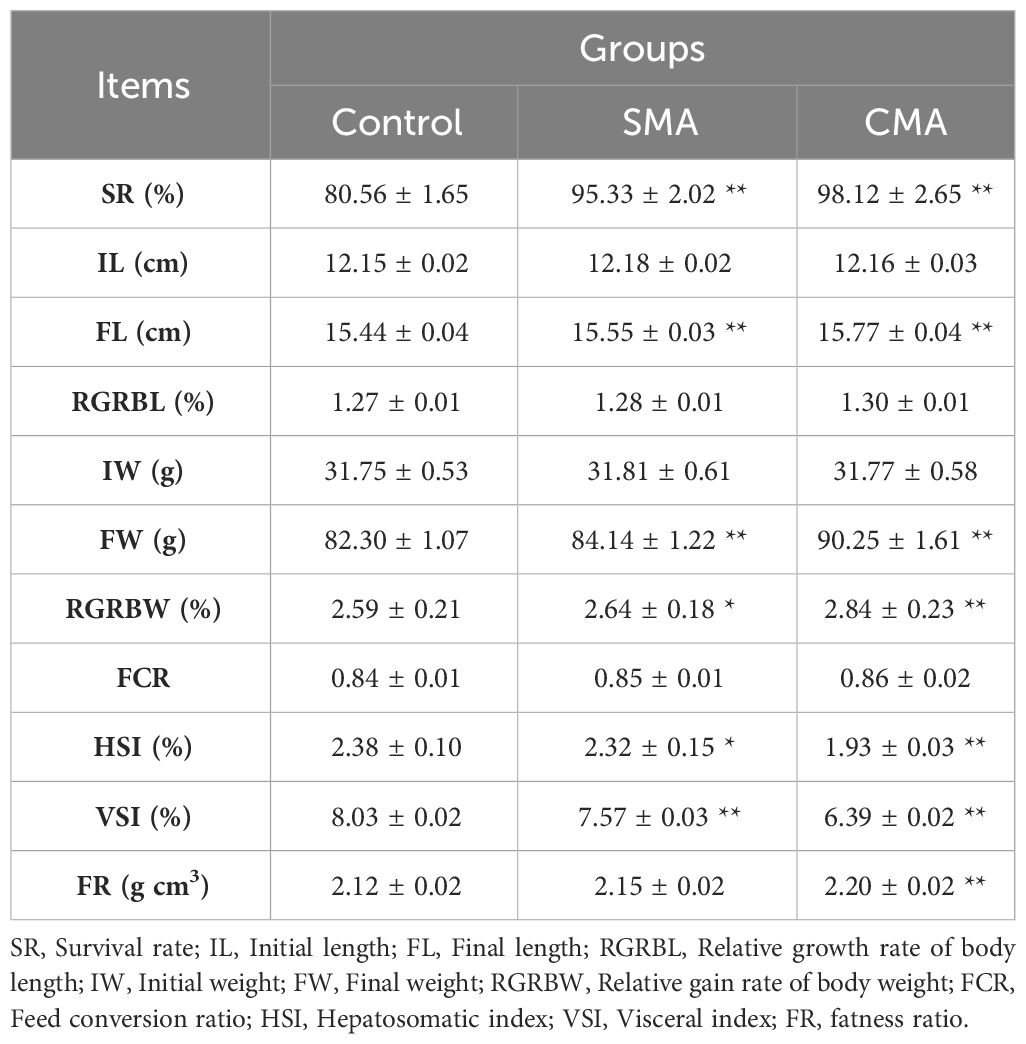
Table 1 Comparison of the growth performance of M. salmoides treated with different experimental feeds. *P ≤ 0.05 and **P ≤ 0.01 were calculated by one-way ANOVA compared with the control.
To investigate the impact of microbial agents on the antioxidant capacity of M. salmoides, we determined the antioxidant enzymatic activity and MDA content in both serum and liver samples at the 41-day feeding time point. The data demonstrated that the activities of SOD, CAT, GSH-Px, whether in serum or liver, were significantly higher in CMA treatment group compared to the control and SMA treatment group. Meanwhile, the MDA content was found to be significantly lower in CMA treated M. salmoides samples than in other treatments, irrespective of whether serum (Figure 4A) or liver (Figure 4B). Additionally, Pearson correlation analysis of the growth indices and antioxidant correlation indicators showed that SOD activity, CAT activity, MDA content, and FR were significantly positively correlated with the growth performance indicators of M. salmoides (Figure 4C).
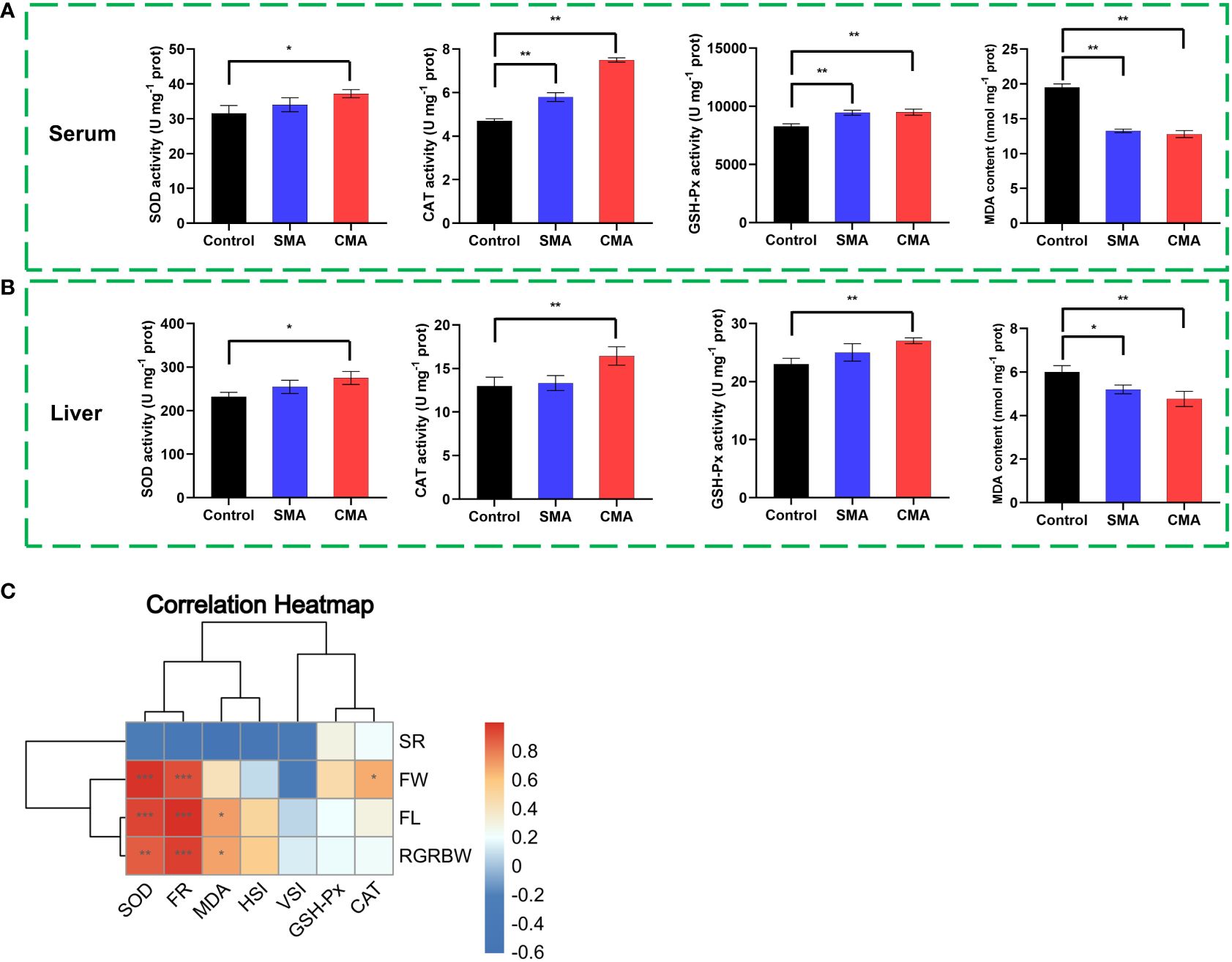
Figure 4 The effects of microbial agents on the growth performance and antioxidant enzyme activity of M. salmoides. The enzymatic activities of SOD, CAT, GSH-Px and the content of MDA in the (A) serum and (B) liver of M. salmoides. (C) The correlation heatmap between growth indices and antioxidant indicators. *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001 were calculated by one-way and two-way ANOVA compared with the control.
3.4 Modification of gut microbiota
In this study, 112645 clean reads and 110818 clean tags per sample were generated from the Illumina MiSeq platform, while the average length of the sequences was calculated to be 456.24 bp per sample. The dilution curves of all samples exhibited a consistently smooth pattern, with a coverage exceeding 99.7%, suggesting that the sequencing depth was sufficient to reflect the biological information concerning the gut microbiota community of M. salmoides (Figure 5).
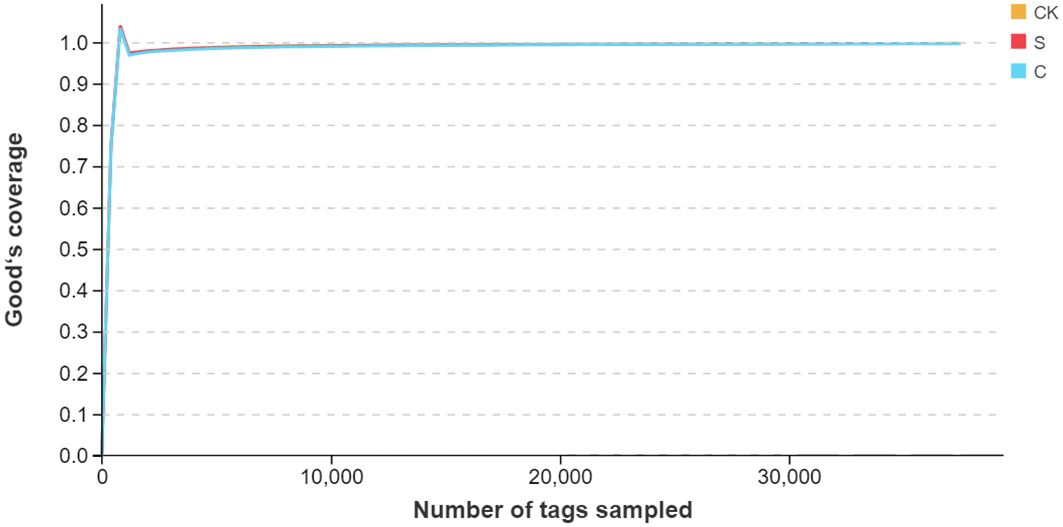
Figure 5 The gut microbiota species dilution curve of each group. The horizontal axis represented the quantity of extracted tags, while the vertical axis represented the calculation of the diversity index value coinciding with the extraction of the respective number of tags.
To evaluate the α-diversity of the gut microbiota of M. salmoides under different treatments, we measured the species richness by calculating the chao1 and ACE indices and the species diversity by calculating the Shannon and Simpson indices. The data showed that compared to the control group, the indices of ACE, chao1, and Shannon were significantly higher in SMA and CMA treatment groups, while Simpson index was the lowest in CMA treated samples (Table 2), indicating that the application of microbial agents, particularly CMA, led to an increase in both species richness and diversity within the gut microbiota of M. salmoides.
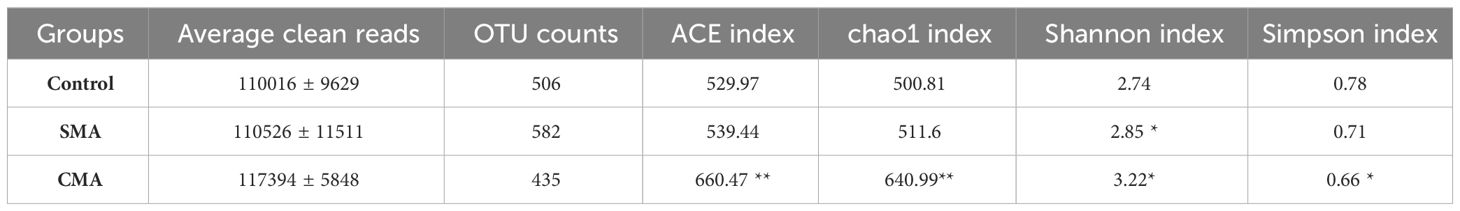
Table 2 Numbers of reads and α-diversity indices of M. salmoides under different treatments. *P ≤ 0.05 and **P ≤ 0.01 were calculated by one-way ANOVA compared with the control.
At the same time, PCoA on OTU level was analyzed to show the β-diversity in the gut microbiota of M. salmoides fed with different diets. As shown in Figure 6A, the community composition of the gut microbiota in M. salmoides differed by different treatments, revealing a variance of 39.58% for PCo1 and 12.73% for PCo2. Meanwhile, to confirm the gut microflora of M. salmoides fed with different diets, the gut microflora composition was shown in the Venn diagram. While 256 OTUs were shared among all three treatments, unique OTU numbering 149, 100, and 206 were identified in the control, SMA, and CMA treatment groups, respectively (Figure 6B).
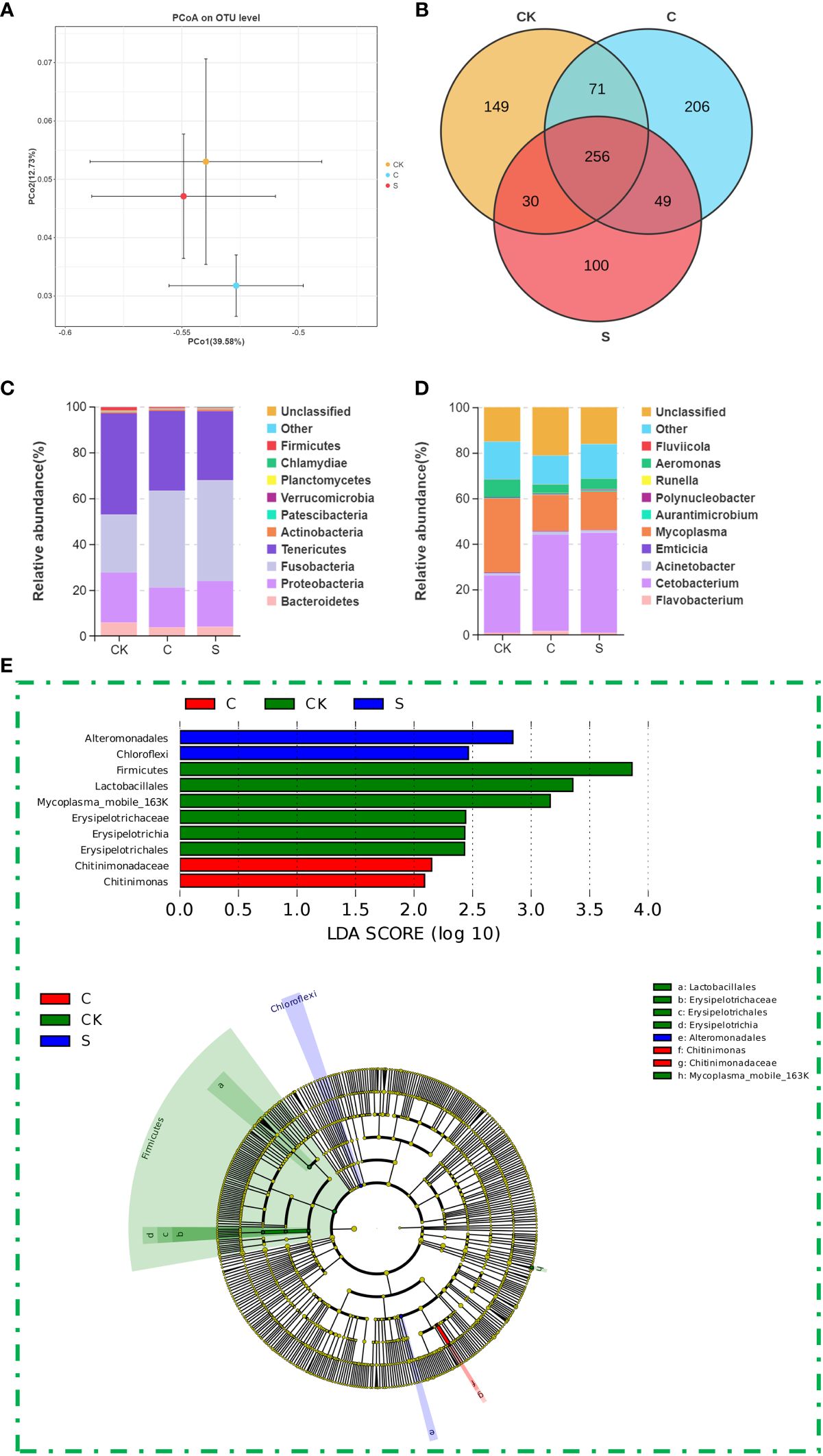
Figure 6 The effects of microbial agents on gut microbiota of M. salmoides. (A) PCoA of the gut microbial composition of M. salmoides under different treatments based on weighted UniFrac distances. (B) The Venn diagram of OTU comparing the unique and shared genera number in gut microbiome of M. salmoides under different treatments. Relative abundance of the top 10 bacterial of M. salmoides under different treatments at (C) phylum level and (D) genus level. (E) The differential flora of M. salmoides under different treatments analyzed by LDA and LEFSe.
Species distribution analysis at the phylum level showed that among the 29 prokaryotic phyla identified from the 16S rRNA gene sequences, Fusobacteria, Tenericutes, and Proteobacteria were the predominant phyla in these treatment groups. A comparison between groups showed that Fusobacteria had a higher relative abundance in SMA treated samples (25.23% in CK, 43.95% in SMA, 42.29% in CMA), while the relative abundance of Tenericutes was higher in CK (44.19% in CK, 30.20% in SMA, 34.73% in CMA), the relative abundance of Proteobacteria was lower in CMA treated samples (21.85% in CK, 19.90% in SMA, 17.30% in CMA) (Figure 6C). Further species distribution analysis was performed at the genus level using all detected bacterial genera. The results showed that Mycoplasma, Cetobacterium, and Aeromonas were the prominent genera among these treatment groups, following by Flavobacterium, Acinetobacter, Emticicia, Polynucleobacter, Aurantimicrobium, Fluviicola, and Runella. Among them, the corresponding dominant bacteria genera were Mycoplasma (32.50%), Cetobacterium (25.23%), Aeromonas (7.69%), and Flavobacterium (0.98%) of the control group, Cetobacterium (43.95%), Mycoplasma (16.96%), Aeromonas (4.72%), and Flavobacterium (0.92%) of SMA group, as well as Cetobacterium (42.29%), Mycoplasma (16.02%), Aeromonas (3.89%), and Flavobacterium (1.67%) of CMA group (Figure 6D). Furthermore, we investigated whether the gut microbes associated with common diet differed from the diet containing microbial agents by using LDA and LEFSe. The gut microbiota with LDA threshold greater than 2.0 were screened, and 10 distinct bacterial groups, characterized by varying classification levels, displayed significant differences. As shown in Figure 6E, the identified biomarkers included Mycoplasma mobile 163K species (Tenericutes phylum), Erysipelotrichaceae family (Firmicutes phylum), and Erysipelotrichales and Lactobacillales orders (Firmicutes phylum) in the control group, Alteromonadales order (Proteobacteria phylum) and Chloroflexi phylum in SMA group, as well as Chitinimonas genus (Proteobacteria phylum) and Chitinimonadaceae family (Proteobacteria phylum) in CMA group.
4 Discussion
The evaluation of water quality serves as a critical factor in assessing the triumph or lack thereof in aquaculture, particularly in intensive aquaculture (Eze et al., 2023). The efficient purification of water quality stands as an essential element for ensuring the long-term viability of intensive aquaculture (Mramba and Kahindi, 2023). Furthermore, water quality plays a crucial role in the cultivation of M. salmoides, affecting not only the physical features and muscular attributes, but also the nutrient composition, mineral elements, and volatile substances within the muscle (Jia et al., 2022). Meanwhile, diminished water quality exerts an influence on the gut microbiota through signaling networks like Nrf2-KEAP1 and MAPK, leading to a decline in both the growth rate and nutritional quality of fish (Subaramaniyam et al., 2023). Therefore, improving water quality is of great significance for the cultivation of M. salmoides. Recent studies have revealed that the presence of microbial agents alters the overall quality of soil by modifying the bacterial community structure and the chemical properties (Du et al., 2022; Ahsan et al., 2023; Wang et al., 2023). The impact of microbial agents on the water quality of aquaculture system has garnered increasing attention from researchers (Hoseinifar et al., 2023). In the present study, compared to the control, CMA treatment delayed the increase of pH (a decline of 2.96%), improved the DO value (a rise of 4.63%), and inhibited the eutrophication of M. salmoides aquaculture system by decreasing levels of ammonia nitrogen (a decline of 41.75%), nitrite (a decline of 19.35%), total phosphorus (a decline of 9.04%), and total nitrogen (a decline of 22.45%) at 7 d. Meanwhile, the water quality of CMA treated group was better than that of SAM treated group, suggesting that the application of microbial agents effectively improved the water quality of M. salmoides aquaculture system, with CMA treatment yielding superior outcomes. It has been proven that the efficacy of CMA in controlling Fusarium oxysporum in banana surpassed that of SMA, with a more distinct strain classification leading to enhanced control effects (Du et al., 2022), corroborating our results.
In addition to improving the water quality, microbial agent treatment produced multiple effects on the bacterial community structure and diversity of aquacultural water systems (Hoseinifar et al., 2023). The presence of microbial agents in water facilitates their utilization of substrates for proliferation while simultaneously exerting a competitive inhibitory effect on the growth of pathogenic bacteria (Sahu et al., 2008). In this study, α-diversity and β-diversity analysis of the microbiota showed that compared to the control and SMA treatment group, CMA treatment group exhibited higher diversity among the microbial community, particularly of chao1 index at 32 d (17.58% higher than that of the control group), indicating the effectiveness of CMA treatment in improving species diversity of water microorganism and regulating the structure of water flora. There was a strong correlation between water quality and the species diversity of water microorganism, contributing to the stability of aquaculture ecosystem for M. salmoides (Nie et al., 2022). Simultaneously, LEFSe analysis demonstrated that the relative abundance of Bacteroides was higher in the control group at 32 d (p value < 0.05), indicating that compared to SMA and CMA treatment group, the aquatic environment of the control group has undergone severe eutrophication (Wagner and Loy, 2002). Additionally, the diversity of microbial community was most prominent in CMA treated samples (Sun et al., 2023), following by SAM (Dutra et al., 2017) and the control group (Liu et al., 2018). Notably, Alpha proteobacteria was consistently highly enriched in CMA treatment group, leading to the suppression of organic pollutants and ammonia oxide of water body (Li et al., 2016).
Beyond water quality, microbial agents alter the gut flora of a host and regulate host immunity and growth through the gut-brain metabolic axis (Berding et al., 2021). The application of Yarrowia lipolytica (189.82 g/kg) significantly increased the relative abundance of Mycoplasma and Enterobacteriacea content, resulting in the increase of the weight gain rate and specific growth rate of juvenile M. salmoides (Fei et al., 2022). In this study, compared to the control group, SMA and CMA treatment increased the diversity of the dominating gut microbiota of M. salmoides, especially CMA treated samples (a rise of 28.13% in ACE, a rise of 27.67% in chao1, a rise of 17.52% in Shannon, and a decline of 15.38% in Simpson). According to the effects of CMA treatment on the survival rate, final length, final weigh, relative gain rate of body weight, hepatosomatic index, visceral index, and fatness ratio, the results of combined analysis suggested that CMA treatment maintained the physiology and homeostasis of M. salmoides by modulating the gut microbial structure. At the same time, through LEFSe analysis, the family Erysipelotrichaceae was more prevalent in the control group, which was positively associated with intestinal inflammation (Yang et al., 2022), suggesting that the M. salmoides samples that were nourished with SMA and CMA exhibited a reduced susceptibility to gastrointestinal inflammation in comparison to the group fed the control diet. Additionally, changes in the composition, diversity, and functional traits of gut microbiota led to increased antioxidant activity in animals (Shandilya et al., 2022). Consequently, this improvement contributes to the maintenance of immune homeostasis and the suppression of chronic inflammation (Vaiserman et al., 2017). In this study, compared to the control and SMA treatment samples, CMA treatment resulted in increased activities of SOD, CAT, and GSH-Px, as well as lower MDA level in both the serum and liver. Besides, it was found that the enhanced antioxidant capacity was positively associated with the growth and health of M. salmoides, suggesting that CMA treatment enhanced the antioxidative capacity through modulating the structure and function of gut microbiota, consequently affecting the development and survival of M. salmoides. The potential mechanism could stem from the enhancement of gut microbiota following CMA treatment, and the pivotal role played by secondary metabolites generated by beneficial bacteria is highlighted as they contribute significantly towards ameliorating oxidative stress and inflammation, thereby improving the antioxidant capacity and inhibiting peroxide production of the host (Wan et al., 2021). Therefore, our findings suggest that CMA treatment resulted in enhanced water quality within the circulating culture system, which contributed to the enrichment of the gut microbial community structure in M. salmoides. Consequently, the proliferation of beneficial bacteria was facilitated, leading to the enhancement of the ROS scavenging system in both the liver and serum, resulting in the improved the survival and development of M. salmoides. In addition, evaluating the field validity of this new approach using M. salmoides and other aquaculture products in natural environment should be considered, which will be one of our future research directions.
5 Conclusion
In summary, we applied a novel type of CMA (yeast, Bacillus subtilis, and lactic acid bacteria) in the recirculating aquaculture system of M. salmoides. The results of water quality parameters showed that CMA treatment decreased the pH level and increased the DO values during the later stage, as well as inhibiting the accumulation of total nitrogen, total phosphorus, ammonia nitrogen, and nitrite during the whole period. The 16s sequencing result of water body demonstrated that CMA treatment improved the species diversity of water microorganism and regulated the structure of water flora, as well as altering the diversity pattern of bacterial related to eutrophication. Subsequently, the improvement of water quality and the change of microflora structure significantly affected the α-diversity and β-diversity of gut microbiota in M. salmoides. This, in turn, hindered the generation and proliferation of harmful bacteria, leading to enhanced antioxidant enzymatic activity and reduced MDA level in both the serum and liver, resulting in an overall improvement in the survival and growth of M. salmoides. This study not only gains insight into the positive role of microbial agents on M. salmoides, but also lays the foundation for CMA application in aquaculture.
Data availability statement
All 16s rRNA sequencing data were deposited in the National Center for Biotechnology Information sequence read archive (SRA) repository, and the BioSample accession numbers 34043908-34043948, inclusive.
Ethics statement
The animal study was approved by Experimental Animal Management Ordinance of Zhejiang Province, China. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
QL: Conceptualization, Investigation, Methodology, Writing – original draft. L-HS: Methodology, Software, Writing – original draft. B-CY: Methodology, Software, Writing – original draft. J-HJ: Methodology, Software, Writing – original draft. J-MC: Investigation, Validation, Writing – original draft. GZ: Investigation, Validation, Writing – original draft. L-MG: Investigation, Validation, Writing – original draft. JP: Investigation, Validation, Writing – original draft. J-LG: Data curation, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by Key research and development project of Zhejiang Province (Grant No. 2021C02024), Huzhou natural science fund project (Grant No. 2022YZ36) and Zhejiang public welfare technology application research program (Grant No. LGF22C01000).
Acknowledgments
The authors thank Prof. Gang Zheng for the assistance with microbial agents.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1379711/full#supplementary-material
References
Ahsan T., Tian P. C., Gao J., Wang C., Liu C., Huang Y. Q. (2023). Effects of microbial agent and microbial fertilizer input on soil microbial community structure and diversity in a peanut continuous cropping system. J. Adv. Res. 23, 00367–00373. doi: 10.1016/j.jare.2023.11.028
Berding K., Vlckova K., Marx W., Schellekens H., Stanton C., Clarke G., et al. (2021). Diet and the microbiota-gut-brain axis: sowing the seeds of good mental health. Adv. Nutr. 12, 1239–1285. doi: 10.1093/advances/nmaa181
Che M., Lu Z., Liu L., Li N., Ren L., Chi S. (2023). Dietary lysophospholipids improves growth performance and hepatic lipid metabolism of largemouth bass (Micropterus salmoides). Anim. Nutr. 13, 426–434. doi: 10.1016/j.aninu.2023.04.007
Chen S. Q., Daly P., Zhou D. M., Li J. J., Wang X. Y., Deng S., et al. (2022). The use of mutant and engineered microbial agents for biological control of plant diseases caused by Pythium: Achievements versus challenges. Fungal Biol. Rev. 40, 76–90. doi: 10.1016/j.fbr.2022.03.001
Chen C., Wu Y., Li J., Wang X., Zeng Z., Xu J., et al. (2023). TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 16, 1733–1742. doi: 10.1016/j.molp.2023.09.010
Dan Z., Mao X., Liu Q., Guo M., Zhuang Y., Liu Z., et al. (2020). Altered gut microbial profile is associated with abnormal metabolism activity of Autism Spectrum Disorder. Gut Microbes 11, 1246–1267. doi: 10.1080/19490976.2020.1747329
Du C., Yang D., Ye Y., Pan L., Zhang J., Jiang S., et al. (2022). Construction of a compound microbial agent for biocontrol against Fusarium wilt of banana. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1066807
Dutra F. M., Rönnau M., Sponchiado D., Forneck S. C., Freire C. A., Ballester E. L. C. (2017). Histological alterations in gills of Macrobrachium amazonicum juveniles exposed to ammonia and nitrite. Aquat Toxicol. 187, 115–123. doi: 10.1016/j.aquatox.2017.04.003
Edwards T. M., Puglis H. J., Kent D. B., Durán J. L., Bradshaw L. M., Farag A. M. (2024). Ammonia and aquatic ecosystems - A review of global sources, biogeochemical cycling, and effects on fish. Sci. Total Environ. 907, 167911. doi: 10.1016/j.scitotenv.2023.167911
El-Saadony M. T., Alagawany M., Patra A. K., Kar I., Tiwari R., Dawood M. A. O., et al. (2021). The functionality of probiotics in aquaculture: An overview. Fish Shellfish Immunol. 117, 36–52. doi: 10.1016/j.fsi.2021.07.007
Eze E., Kirby S., Attridge J., Ajmal T. (2023). Aquaculture 4.0: hybrid neural network multivariate water quality parameters forecasting model. Sci. Rep. 13, 16129. doi: 10.1038/s41598-023-41602-7
Fei H., Cheng Y., Zhang H., Yu X., Yi S., Huang M., et al. (2022). Effect of Autolyzed Yarrowia lipolytica on the Growth Performance, Antioxidant Capacity, Intestinal Histology, Microbiota, and Transcriptome Profile of Juvenile Largemouth Bass (Micropterus salmoides). Int. J. Mol. Sci. 23, 10780. doi: 10.3390/ijms231810780
Giri S. S., Kim H. J., Kim S. G., Kim S. W., Kwon J., Lee S. B., et al. (2020). Immunomodulatory role of microbial surfactants, with special emphasis on fish. Int. J. Mol. Sci. 21, 7004. doi: 10.3390/ijms21197004
Guo J. L., Kuang W. M., Zhong Y. F., Zhou Y. L., Chen Y. J., Lin S. M. (2020). Effects of supplemental dietary bile acids on growth, liver function and immunity of juvenile largemouth bass (Micropterus salmoides) fed high-starch diet. Fish Shellfish Immunol. 97, 602–607. doi: 10.1016/j.fsi.2019.12.087
Heikema A. P., Horst-Kreft D., Boers S. A., Jansen R., Hiltemann S. D., de Koning W., et al. (2020). Comparison of Illumina versus Nanopore 16S rRNA Gene Sequencing of the Human Nasal Microbiota. Genes 11, 1105. doi: 10.3390/genes11091105
Hoseinifar S. H., Maradonna F., Faheem M., Harikrishnan R., Devi G., Ringø E., et al. (2023). Sustainable ornamental fish aquaculture: the implication of microbial feed additives. Animals 13, 1583. doi: 10.3390/ani13101583
Hugerth L. W., Wefer H. A., Lundin S., Jakobsson H. E., Lindberg M., Rodin S., et al. (2014). DegePrime, a program for degenerate primer design for broad-taxonomic-range PCR in microbial ecology studies. Appl. Environ. Microbiol. 80, 5116–5123. doi: 10.1128/AEM.01403-14
Jia S. P., Wang L., Zhang J. M., Zhang L., Ma F. R., Huang M. L., et al. (2022). Comparative study on the morphological characteristics and nutritional quality of largemouth bass (Micropterus salmoides) cultured in an aquaculture system using land-based container with recycling water and a traditional pond system. Aquaculture 549, 737721. doi: 10.1016/j.aquaculture.2021.737721
Johnson J. S., Spakowicz D. J., Hong B. Y., Petersen L. M., Demkowicz P., Chen L., et al. (2019). Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10, 5029. doi: 10.1038/s41467-019-13036-1
Julian P., Gu B. (2015). Mercury accumulation in largemouth bass (Micropterus salmoides Lacépède) within marsh ecosystems of the Florida Everglades, USA. Ecotoxicology 24, 202–214. doi: 10.1007/s10646-014-1373-9
Li W., Wang F., Zhang J., Qiao Y., Xu C., Liu Y., et al. (2016). Community shift of biofilms developed in a full-scale drinking water distribution system switching from different water sources. Sci. Total Environ. 544, 499–506. doi: 10.1016/j.scitotenv.2015.11.121
Liang J., Jin Y., Wen X., Mi J., Wu Y. (2020). Adding a complex microbial agent twice to the composting of laying-hen manure promoted doxycycline degradation with a low risk on spreading tetracycline resistance genes. Environ. pollut. 265, 114202. doi: 10.1016/j.envpol.2020.114202
Liu Z., Qiuqian L., Yao Z., Wang X., Huang L., Zheng J., et al. (2018). Effects of a commercial microbial agent on the bacterial communities in shrimp culture system. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02430
Maciag T., Kozieł E., Rusin P., Otulak-Kozieł K., Jafra S., Czajkowski R. (2023). Microbial Consortia for Plant Protection against Diseases: More than the Sum of Its Parts. Int. J. Mol. Sci. 24, 12227. doi: 10.3390/ijms241512227
Mramba R. P., Kahindi E. J. (2023). Pond water quality and its relation to fish yield and disease occurrence in small-scale aquaculture in arid areas. Heliyon 9, e16753. doi: 10.1016/j.heliyon.2023.e16753
Narayanan M., Kandasamy S., Lee J., Barathi S. (2023). Microbial degradation and transformation of PPCPs in aquatic environment: A review. Heliyon 9, e18426. doi: 10.1016/j.heliyon.2023.e18426
Ni J., Tokuda G. (2013). Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol. Adv. 31, 838–850. doi: 10.1016/j.biotechadv.2013.04.005
Nie Z., Zheng Z., Zhu H., Sun Y., Gao J., Gao J., et al. (2022). Effects of submerged macrophytes (Elodea nuttallii) on water quality and microbial communities of largemouth bass (Micropterus salmoides) ponds. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1050699
Niimi A. J., Beamish F. W. H. (1974). Bioenergetics and growth of largemouth bass (Micropterus salmoides) in relation to body weight and temperature. Can. J. Zool 52, 447–456. doi: 10.1139/z74-056
Øverland M., Skrede A. (2017). Yeast derived from lignocellulosic biomass as a sustainable feed resource for use in aquaculture. J. Sci. Food Agric. 97, 733–742. doi: 10.1002/jsfa.8007
Peng Z., Zhang C., Yin B., He Y., Li W., Wang J., et al. (2023). TRIM21 of Micropterus salmoides exerts antiviral roles against largemouth bass ulcer syndrome virus. Fish Shellfish Immunol. 142, 109176. doi: 10.1016/j.fsi.2023.109176
Ren W., Wu H., Guo C., Xue B., Long H., Zhang X., et al. (2021). Multi-strain tropical bacillus spp. as a potential probiotic biocontrol agent for large-scale enhancement of mariculture water quality. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.699378
Sahu M. K., Swarnakumar N. S., Sivakumar K., Thangaradjou T., Kannan L. (2008). Probiotics in aquaculture: importance and future perspectives. Indian J. Microbiol. 48, 299–308. doi: 10.1007/s12088-008-0024-3
Shandilya S., Kumar S., Kumar Jha N., Kumar Kesari K., Ruokolainen J. (2022). Interplay of gut microbiota and oxidative stress: Perspective on neurodegeneration and neuroprotection. J. Adv. Res. 38, 223–244. doi: 10.1016/j.jare.2021.09.005
Subaramaniyam U., Allimuthu R. S., Vappu S., Ramalingam D., Balan R., Paital B., et al. (2023). Effects of microplastics, pesticides and nano-materials on fish health, oxidative stress and antioxidant defense mechanism. Front. Physiol. 14. doi: 10.3389/fphys.2023.1217666
Sun D., Xie B., Li J., Huang X., Chen J., Zhang F. (2023). A low-cost microbial fuel cell based sensor for in-situ monitoring of dissolved oxygen for over half a year. Biosens Bioelectron 220, 114888. doi: 10.1016/j.bios.2022.114888
Vaiserman A. M., Koliada A. K., Marotta F. (2017). Gut microbiota: A player in aging and a target for anti-aging intervention. Ageing Res. Rev. 35, 36–45. doi: 10.1016/j.arr.2017.01.001
Valencia-Castañeda G., Frías-Espericueta M. G., Vanegas-Pérez R. C., Chávez-Sánchez M. C., Páez-Osuna F. (2019). Toxicity of ammonia, nitrite and nitrate to Litopenaeus vannamei juveniles in low-salinity water in single and ternary exposure experiments and their environmental implications. Environ. Toxicol. Pharmacol. 70, 103193. doi: 10.1016/j.etap.2019.05.002
Wagner M., Loy A. (2002). Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol. 13, 218–227. doi: 10.1016/S0958-1669(02)00315-4
Wan M. L. Y., Co V. A., El-Nezami H. (2021). Dietary polyphenol impact on gut health and microbiota. Crit. Rev. Food Sci. Nutr. 61, 690–711. doi: 10.1080/10408398.2020.1744512
Wang Q., Song J., Zhang J., Qin X., Kang Y., Huang S., et al. (2023). Effects of microbial agent application on the bacterial community in ginger rhizosphere soil under different planting years. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1203796
Yang H., Wu J., Huang X., Zhou Y., Zhang Y., Liu M., et al. (2022). ABO genotype alters the gut microbiota by regulating GalNAc levels in pigs. Nature 606, 358–367. doi: 10.1038/s41586-022-04769-z
Yang J., Zhang X., Xie Y., Song C., Sun J., Zhang Y., et al. (2017). Ecogenomics of zooplankton community reveals ecological threshold of ammonia nitrogen. Environ. Sci. Technol. 51, 3057–3064. doi: 10.1021/acs.est.6b05606
Yin Y., Chen X., Gui Y., Zou J., Wang Q., Qiu L., et al. (2022). Risk and benefit assessment of potential neurodevelopment effect resulting from consumption of cultured largemouth bass (Micropterus salmoides) in China. Environ. Sci. pollut. Res. Int. 29, 89788–89795. doi: 10.1007/s11356-022-22065-1
Zehra A., Raytekar N. A., Meena M., Swapnil P. (2021). Efficiency of microbial bio-agents as elicitors in plant defense mechanism under biotic stress: A review. Curre Res. Micro Sci. 2, 100054. doi: 10.1016/j.crmicr.2021.100054
Zhai J. X., Jiumu L. M., Cong L., Wu Y. N., Dai L. Y., Zhang Z. M., et al. (2020). Reed decomposition under Bacillus subtilis addition conditions and the influence on water quality. Ecohydrol Hydrobiol 20, 504–512. doi: 10.1016/j.ecohyd.2019.11.003
Zhang Y., Zhang H., Liu Q., Duan L., Zhou Q. (2023). Total nitrogen and community turnover determine phosphorus use efficiency of phytoplankton along nutrient gradients in plateau lakes. J. Environ. Sci. (China) 124, 699–711. doi: 10.1016/j.jes.2022.02.005
Keywords: Micropterus salmoides, microbial agents, water quality, gut microbiota, 16S rRNA sequencing
Citation: Li Q, Sun L-H, Yang B-C, Jiang J-H, Chen J-M, Zheng G, Gao L-M, Peng J and Guo J-L (2024) Effectiveness of a novel compound microbial agent for water quality and the gut microbiota of Micropterus salmoides. Front. Mar. Sci. 11:1379711. doi: 10.3389/fmars.2024.1379711
Received: 08 February 2024; Accepted: 02 April 2024;
Published: 03 May 2024.
Edited by:
Marcel Martinez-Porchas, National Council of Science and Technology (CONACYT), MexicoReviewed by:
Vikash Kumar, Central Inland Fisheries Research Institute (ICAR), IndiaD. K. Meena, Central Inland Fisheries Research Institute (ICAR), India
Copyright © 2024 Li, Sun, Yang, Jiang, Chen, Zheng, Gao, Peng and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Lin Guo, wavegjl@aliyun.com
 Qian Li
Qian Li Li-Hui Sun1,2,3
Li-Hui Sun1,2,3  Jian-Lin Guo
Jian-Lin Guo