Effect of coastal hypoxia on bacterial diversity as elucidated through 16S rRNA amplicon sequencing
- 1CSIR - National Institute of Oceanography, Biological Oceanography Division, Goa, India
- 2School of Earth, Ocean and Atmospheric Sciences, Goa University, Goa, India
- 3CSIR - National Institute of Oceanography, Chemical Oceanography Division, Goa, India
The formation of oxygen-depleted zones in the bottom waters is one of the most widespread phenomena in coastal areas. Upwelling episodes along India’s west coast due to the southwest monsoon increase biological productivity, further lowering the dissolved oxygen in the upwelled waters, which intensifies annually between June and October. Here, we have determined the changes in the microbial community in response to the varying oxygen levels and other physicochemical parameters at the Candolim Time Series Station using high-throughput sequencing. Amplicon Sequence Variants across all the samples collected in different seasons were mostly affiliated to the phyla Proteobacteria, Actinobacteria, Bacteroidetes, Verrucomicrobia, Chloroflexi, Firmicutes and Planctomycetes, with the most dominant being Proteobacteria (21-41%). Statistical analysis revealed that microbial diversity differed significantly with changing DO, ammonia, nitrate and nitrite concentrations during different seasons. The microbial community shift due to seasonal hypoxia results in the differential biogeochemical cycling of essential nutrients, with certain years seeing redox conditions up to sulphate reduction while certain years seeing only nitrogen loss. Future global warming scenarios will serve as a big challenge for understanding the role of microbial diversity and its implications in the cycling of natural elements.
1 Introduction
Coastal ecosystems play a pivotal role in regulating the global biogeochemical cycles. Some major processes in these ecosystems include the exchange of nutrients, carbon in the form of dissolved organic matter, reactive organic and inorganic trace gases, and various other physical and biogeochemical elements. Coastal ecosystems are significant as they are highly productive areas (Wiggert et al., 2005). The increased biological productivity due to monsoonal upwelling leads to increased organic matter production utilized by heterotrophic microorganisms. The intense decomposition of organic matter leads to rapid utilization of in situ oxygen, leading to its depletion and forming hypoxic conditions in sub-surface waters (Naqvi et al., 2000; Gomes et al., 2019).
The west coast of India experiences coastal hypoxia due to upwelling, which is mainly influenced by the southward movement of the West Indian coastal current during the South West Monsoon (SWM) (Madhupratap et al., 1996). The upwelled waters are already hypoxic (<2 mL L-1) as these are from the thermocline region off the continental shelf of India (Naqvi et al., 2000). During the SWM, the bottom waters having low oxygen levels profoundly impact the biogeochemistry and ecological functioning of the biome and the underlying sediments. Hypoxia can also be caused by the anthropogenic inputs of nutrients and organic matter, leading to a drastic decrease in DO due to eutrophication (Rabalais et al., 2002). Such low oxygen levels threaten marine ecosystems and are also responsible for disturbing benthic microbial habitats. In addition, prolonged hypoxic conditions may change the microbial community in response to oxygen depletion. In a case study carried out in the Black Sea, the presence or absence of oxygen in the water column has been suggested as the main reason for variations in microbial community structure and function (Thamdrup et al., 2000). Beman and Carolan (2013) reported a non-linear relationship between DO concentration and bacterial richness in the oxygen-deficient waters of the Eastern Tropical North Pacific Ocean.
Hypoxic events are often considered one of the major global environmental problems (Diaz and Rosenberg, 2008; Naqvi et al., 2010; Breitburg et al., 2018). The seasonal coastal hypoxic zone of the Eastern Arabian Sea is by far the largest of all the known coastal hypoxic systems formed due to natural and anthropogenic activities (Naqvi et al., 2006; Sudheesh et al., 2020). It is observed that if hypoxic conditions persist for a long time, the organic matter and nutrients tend to accumulate in the sediments resulting in the expansion of the hypoxic zone. This leads to a further decrease in DO levels, thus establishing anoxic conditions, favoring the release of H2S by microorganisms (Diaz and Rosenberg, 2008). The progression of seasonal coastal hypoxia has been shown to follow a predictable pattern that starts with SWM’s advent (June to September). Oxygen depletion might also begin in April-May and gradually intensify with time (Naqvi et al., 2006). The CaTS-G5 station on the West coast of India has been extensively monitored since 1997 to study the changes occurring due to coastal hypoxia. This could be considered a representative station to understand the oxygen deficiency in the coastal regions (Naqvi et al., 2006). The oxygen-deficient conditions intensify towards the end of the southwest monsoon, whereas the water column remains well-oxygenated during other periods.
Prolonged oxygen-deficient episodes often change the bacterial community composition, affecting the ecosystem’s functioning (Reed and Martiny, 2013). As oxygen is the most favorable electron acceptor in aerobic aquatic systems, its availability could influence gene expression of the bacterial community and affect respiratory metabolism. A shift in bacterial community composition during the development of hypoxia was reported by Beman and Carolan (2013) in the oxygen minimum zones of the Eastern North Pacific Ocean, where they recorded a non-linear relationship between bacterial richness and DO concentration. On the contrary, a negative correlation between bacterial richness and DO was reported by Spietz et al., 2015 in a hypoxic estuary. Various bacterial lineages belonging to Alphaproteobacteria, Gammaproteobacteria and Deltaproteobacteria have been reported to be abundant in oxygen-deficient marine waters. Microbes reported from oxygen-depleted waters falling into these lineages are considered to be chemolithoautotrophic sulphur oxidizers that play a crucial role in sulfur and carbon cycling in the ocean ecosystem (Lipsewers et al., 2017). Previous studies, carried out in the Northern Indian Ocean oxygen minimum zone (OMZ), based on metagenomic analysis, reported taxa belonging to Proteobacteria, Actinobacteria, Nitrospinota, Planctomycetes and SAR406 clade; however, the distribution varied along depths and sites sampled (Bandekar et al., 2018; Fernandes et al., 2019). High throughput sequencing techniques and their applications have helped unravel the microbial diversity in environmental samples (Caporaso et al., 2012; Lynch and Neufeld, 2015).
The present study was conducted to investigate the effect of hypoxia on the microbial diversity at the CaTS-G5 station in the Arabian Sea. The microbial communities were deduced by 16S rRNA amplicon sequencing from water samples collected during different seasons, i.e. peak monsoon (July 2018), late southwest monsoon (Sept. 2018) and fall inter-monsoon (Oct. 2018, 2019). The annual variation with respect to anoxia in the bacterial diversity was studied, along with the distribution of bacterial communities in response to the physicochemical environmental variables and predictive functional analysis of the microbial communities.
2 Materials and methods
2.1 Sampling locations and sampling methodology
Water samples were collected at the CaTS-G5 (15.50877° N, 73.6742°E) station along the CaTS transect in the coastal waters of the Eastern Arabian Sea during the following cruises and field trips: SSD052 ORV Sindhu Sadhana (05-07-2018), SSD057 ORV Sindhu Sadhana (20-09-2018), CaTS 157 field trip (04-10-2018) and CaTS 161 field trip (15-10-2019).
Samples were collected from 2 depths based on the oxygen profile, mid-depth and near bottom as 12 m (GV1) and 24 m (GV2) on SSD052, 13 m (GV3) and 25 m (GV4) on SSD057, 18 m (GV5) and 27 m (GV6) on CaTS 157, 18 m (GV7) and 27 m (GV8) on CaTS 161. On board, the ship, the temperature data of the seawater samples were obtained using a Conductivity-Temperature-Depth (Seabird Electronics SBE911) rosette system fitted Niskin bottles. During the field trips, it was obtained using a portable CTD (SeaBird Scientific SBE25 plus V2), and water samples were collected using 5-litre Niskin samplers. The DO samples were analyzed following the Carpenter modification of the Winkler method (Carpenter, 1965). The titration was automated using ODF PC software compiled in LabView, in which the endpoint is UVC, detected photometrically at 365 nm. The nutrients (nitrate, nitrite, ammonium) were analyzed by the Skalar Autoanalyzer. For DNA extraction, ten liters of seawater was filtered through 0.22 µm Sterivex filters (Millipore, United States) using a peristaltic pump (Cole-Parmer India). The Sterivex filter cartridges were filled with DNA storage buffer (50 mM Tris pH 8.3, 40 mM EDTA, and 0.75 M sucrose) and stored at −80°C until further analysis.
2.2 DNA extraction and sequencing
The Sterivex filter cartridge was cracked open aseptically using a clean plyer. The filter paper was cut with a sterile blade and added to a disruptor tube containing SLX Mlus buffer, and DNA extraction was carried out as per the manufacturer’s protocol (OMEGA BioTek, USA). The vacuum-dried DNA samples were outsourced for sequencing to Eurofins Genomics (India). The QC-passed samples were processed for amplicon generation. The amplicon libraries were prepared for 2 x 300 bp chemistry using Nextera XT Index Kit (Illumina Inc.) as per the 16S Metagenomic Sequencing Library preparation protocol. Primers for the amplification of the bacterial 16S V3-V4 region were designed and synthesized at Eurofins Genomics Lab. The primers used were 16S rRNA F GCCTACGGGNGGCWGCA G 16S rRNA R ACTACHVGGGTATCTAA TCC. The amplicon libraries were purified by AMPure XP beads and quantified using a Qubit Fluorometer. Quantity and quality check (QC) of the library was done using Agilent 4200 Tape Station (Agilent Technologies) using D1000 Screen tape as per manufacturer instructions. The QC passed amplicons with the Illumina adaptor were amplified using the primers that add multiplexing index sequences as well as common adapters required for cluster generation as per the standard Illumina protocol.
2.3 Sequence analysis
High-quality clean reads obtained from Illumina were selected using Trimmomatic v 0.38 (Bolger et al., 2014), and the paired-end data was stitched into single-end reads using FLASH (v1.2.11) (Magoč and Salzberg, 2011). ASVs were generated using the DADA2 plugin in the QIIME2 pipeline (Caporaso et al., 2012), and the taxonomic identity was assigned using the Silva database (version 13_8) (McDonald et al., 2012). The resulting biome file was used for further analysis. Abundance estimation, diversity analysis, and rarefaction analysis were carried out using the QIIME program. The sequence data of the 16S rRNA gene was submitted to the National Center for Biotechnology Information (PRJNA 706932).
2.4 Statistical analyses
The variability in the bacterial classes was compared with five environmental variables, temperature, DO, nitrate, nitrite and ammonia, using Canonical Correspondence Analysis (CCA) using PAST-4 software V4.03 (Hammer et al., 2021). QIIME2 pipeline was used for diversity analysis. Beta diversity at the phyla level between the depths and stations was calculated using multivariate scaling analysis (MDS) based on Bray-Curtis distance using Primer 6 software (Clarke et al., 2008). The PCoA analysis was carried out, and plots were obtained using Unweighted and weighted distance metrics in QIIME2.
2.5 Predictive functional analysis
Predictive functional analysis of the microbial communities present in all eight samples was done using Tax4fun2 (Wemheuer et al., 2020). The 16S rRNA sequences were compared with reference sequences in BLAST using the runRefBlast function. The predictive functions were calculated using the Functional Prediction function based on KEGG ortholog reference profiles. Heatmap and statistical analysis representing the functional predictions within the samples was done using the STAMP V2.1.3 software (Parks et al., 2014). Tukey-Kramer test was carried out to check the differences in functional gene composition within the samples.
3 Results
3.1 Physico-chemical characteristics of the water samples
The temperature of the water samples during the study period varied between 21.8 and 32°C, whereas salinity varied between 33.83 and 35.78 PSU. The temperature at the bottom depths was lower than the upper depths during the months from July to Oct 2018, whereas no significant differences were observed in the salinity (Table 1). The DO levels showed a drastic change from July to October 2018, ranging from oxic conditions (2.3 - 4.4 mL L-1) in July 2018, followed by a hypoxic range of 0.1 - 0.5 mL L-1 in September 2018, progressing towards suboxic conditions (<0.1 mL L-1) in upper depths and completely anoxic conditions in the near bottom during October 2018. The near-bottom depths experienced higher oxygen deficiency as compared to the mid-depths. In October 2018, the water samples collected from mid and near-bottom depths exhibited anoxic conditions with DO levels in the range of 0 - 0.2 mL L-1. The nutrient concentrations also showed a seasonal variation, like the DO values. In general, the nitrate concentrations varied between 0.1 and 8.3 µM, nitrite between undetectable levels and 1.6 µM and ammonia between 0.3 and 9.6 µM (Table 1). In 2018, higher nitrate concentrations (3.1 – 6.9 µM) were observed in September compared to other months. High nitrate was also observed in October 2019. In contrast to nitrate, higher ammonium concentrations (6.13 - 7.06 µM) were observed during October 2018, accompanied by low nitrate values (0.01 - 0.06 µM) and the absence of nitrite. High ammonium concentration (9.6 µM) was also observed in the near-bottom waters during July 2018. H2S (1.66 µM) was detected at the lower depth, indicating anoxic conditions in October 2018 (Table 1).
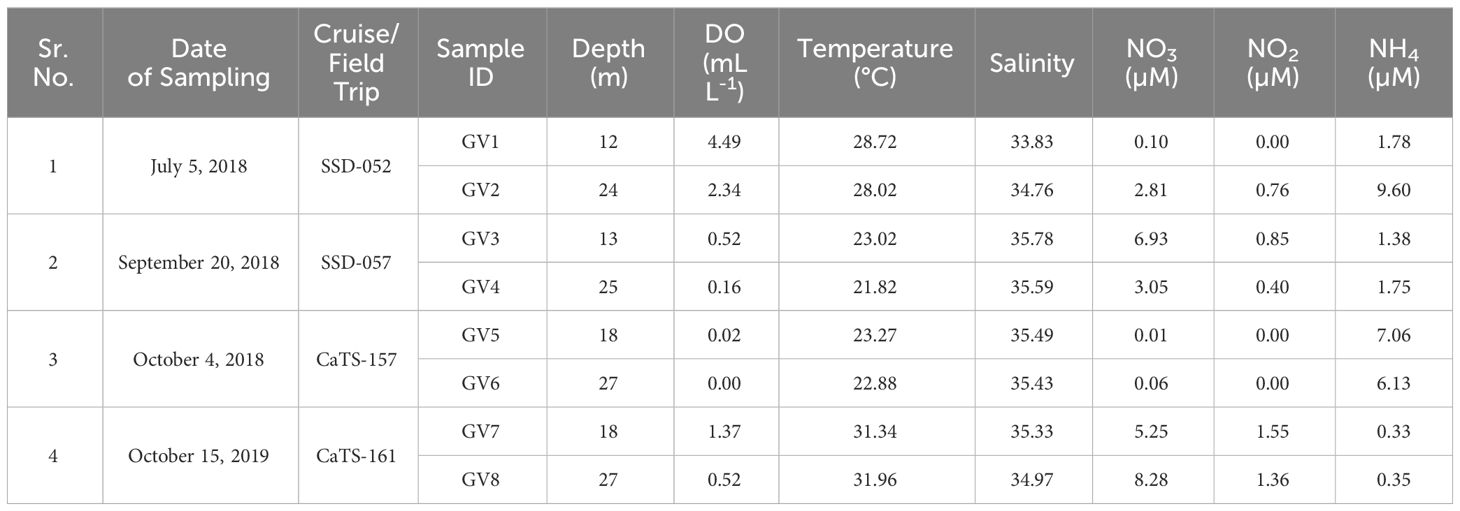
Table 1 Physico-chemical parameters at Candolim Time series Station (CaTS) during the cruise and field trip.
An annual comparison of the physicochemical parameters between October 2018 and 2019 sampling showed that oxygen deficiency intensity was higher in 2018, with oxygen concentration falling in the anoxic range compared to the conditions in 2019 (Table 1). Although in 2019, the sampling site did not experience anoxia, the ammonium concentrations were low (0.33 - 0.35 µM) compared to the observed values in the previous year. The total organic carbon (TOC) was also considerably high during October 2018 in comparison to 2019 (Shetye et al., 2022), with the highest value near the bottom depth, where the conditions were anoxic.
3.2 Prokaryotic community structure
A total of 28 bacterial phyla were identified across all the seasons sampled. The maximum number of ASVs were affiliated with three major bacterial phylogenetic groups, Proteobacteria, Actinobacteria, and Bacteroidota (Figure 1). Proteobacteria dominated across the mid-depth and near-bottom zones accounting for about 42.69 - 68.78% in almost all the seasons, except October 2018, where the mid-depths were dominated by the phylum Actinobacteria (33.2%). Bacteroidota showed a high abundance during September 2018 at both depths; however, it was less abundant during all other seasons. The phyla Planctomycetes and Verrucomicrobiota were evenly distributed across all the seasons at mid and near- bottom depths. Two phyla, Spirochaetes and Nitrospirae, were abundant at lower depths in all the samples. The members of SAR406, Marinimicrobia phyla, were present in all the samples.
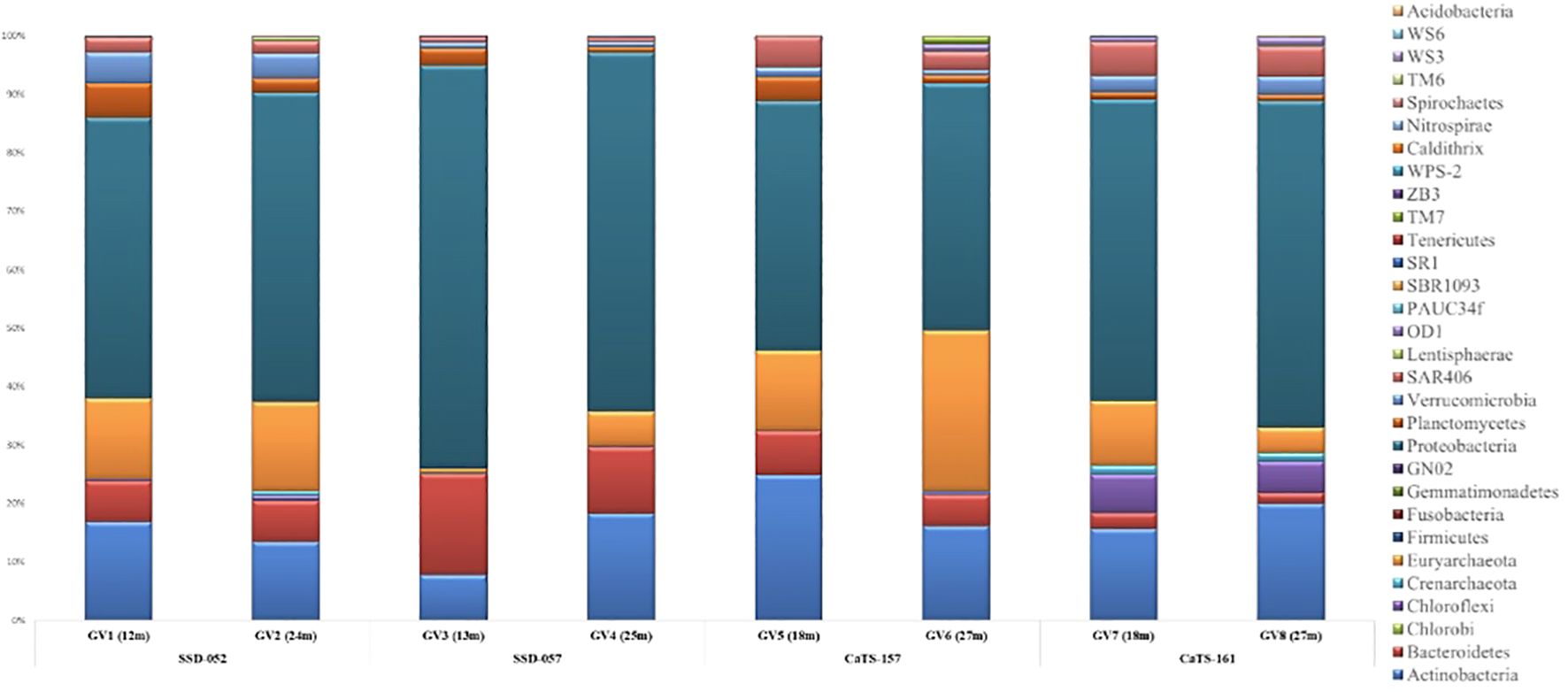
Figure 1 Percentages of relative abundance (Y-axis) and sampling cruise/field trip and depth-wise representation of phyla (X-axis) at the Candolim Time-series station (CaTS) G5.
Among the archaeal phyla, Thermoplasmatota showed dominance during July and October 2018 at both depths. However, during October 2019, Thermoplasmatota showed high abundance in the mid-depths. The two euryarchaeal groups present in all the samples belonged to marine group II and marine group III, with the former being dominant in all the samples. In addition, archaeal phyla Crenarchaeota and Nanoarcheota were present in most samples, albeit not so dominant.
A comparative analysis of relative abundance at the class level between mid and near-bottom depths across all the seasons highlighted that the class Acidobacteria belonging to phyla Actinobacteria showed high abundance in all samples. Among the Proteobacteria, the major classes were Alphaproteobacteria followed by Gammaproteobacteria (Figure 2). It was noted that Alphaproteobacteria were abundant in mid-depth compared to the near-bottom depth in all the samples. Thermoplasmata belonging to the Phylum Thermoplasmatota showed higher abundance in almost all samples except GV3, GV4 and GV8.
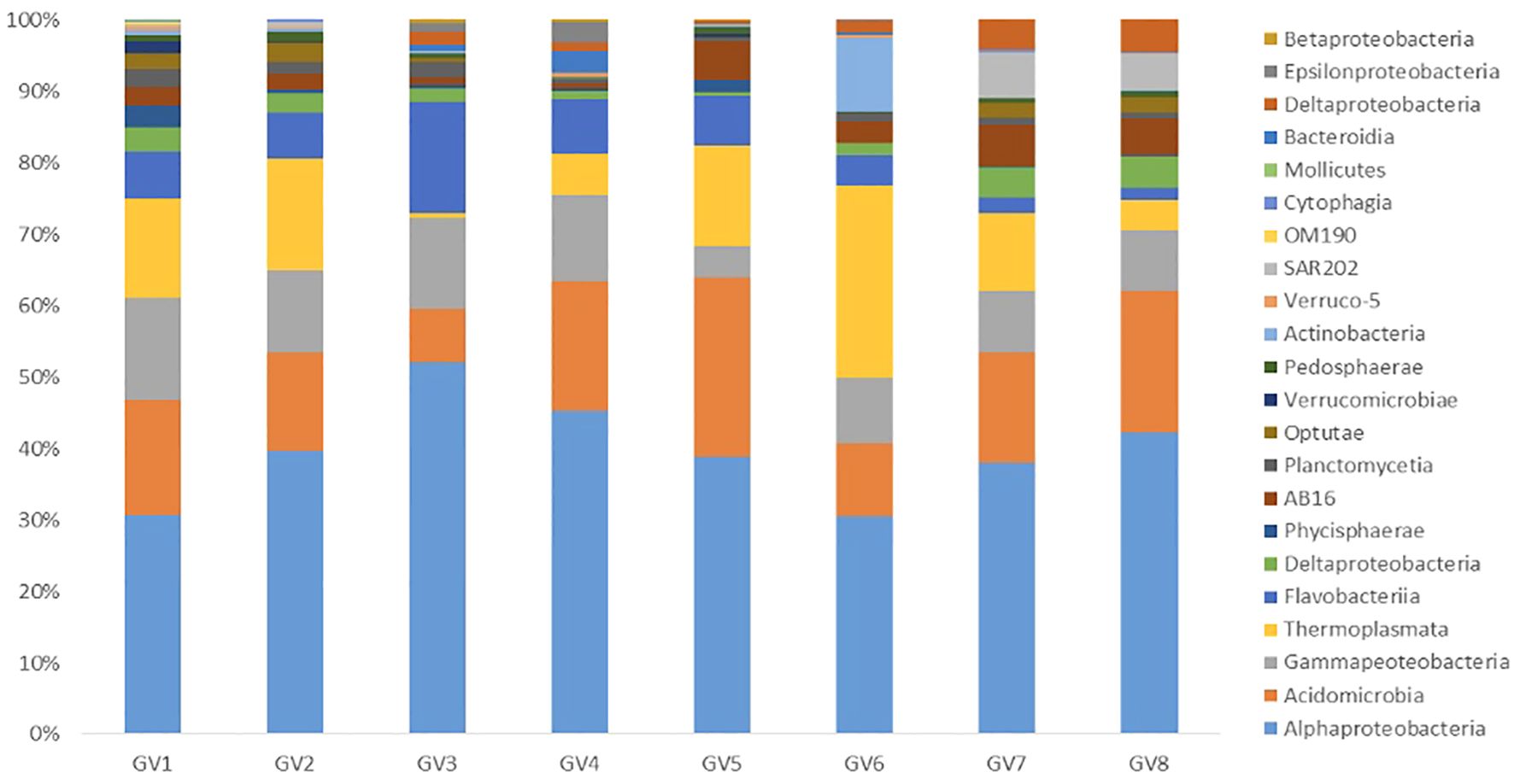
Figure 2 Percentages of relative abundance (Y-axis) and sampling cruise/field trip and depth-wise representation of class (X-axis) at the Candolim Time-series station (CaTS) G5.
At the genus level, Candidatus Actinomarina, NS4 Marine group, NS5 Marine group, Marinimicrobia, clade 1a, AEGEAN-169 marine group, Marine group III, Sva0996 were ubiquitously present in all the samples. In addition, the SUP05 clade was also present in almost all the samples except GV2.
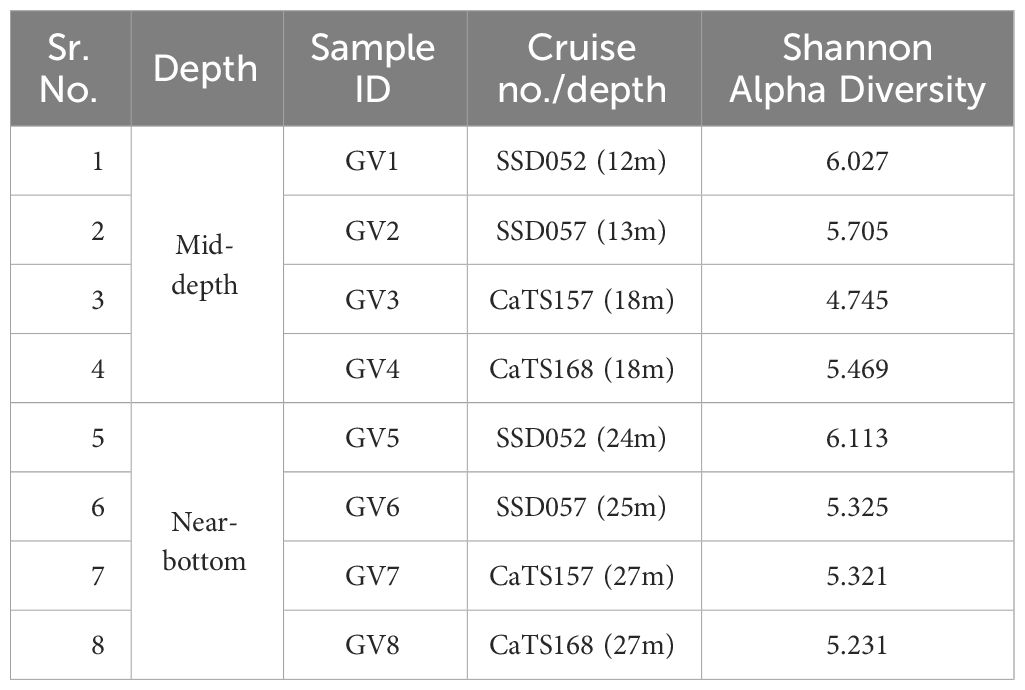
A comparison between the mid-depth samples GV1, GV3, GV5 and GV7 showed a similar trend. Proteobacteria was the dominant phylum in almost all samples except for GV5, irrespective of sampling time. The Shannon alpha diversity analysis of surface depths, i.e., GV1, GV3, GV5 and GV7, indicated that GV3 exhibits higher alpha diversity followed by GV1, GV7 and GV5 (Table 2). Similarly, in the lower depths, Shannon alpha diversity analysis revealed that GV6 displayed higher diversity, followed by GV2, GV4 and GV8. MDS and PCoA analysis showed distinct grouping at phyla level at mid-depth and near bottom depths.
3.3 Correlation between physicochemical properties and the microbial community structure
The effect of environmental variables on microbial community structure at the genus level was studied at the G5 station along both depths. Seasonal and annual variations in community structure with respect to 5 environmental variables were investigated using Canonical Correspondence Analysis (CCA). The triplot obtained reveals a strong positive correlation between dissolved oxygen, ammonia and the genus NS4_marine group, Clade_II and SAR 116 clade belonging to the class Alphaproteobacteria (Figure 3). Archaeal marine group II also shows a slight positive correlation with these two parameters. This correlation was observed in samples GV1 and GV2. NS5 marine group, Marinobacterium, clade 1a, PAUC34f and bacteroidetes showed a positive correlation with nitrate and a negative correlation to temperature. Candidatus nitrosopelagicus, SAR202 clade, Marinimicrobia, SAR324, and AEGEAN-169 marine group were all negatively correlated to nitrite concentration. Among all the environmental parameters analysed, DO and ammonia significantly contributed to the variation in the community in the study area.
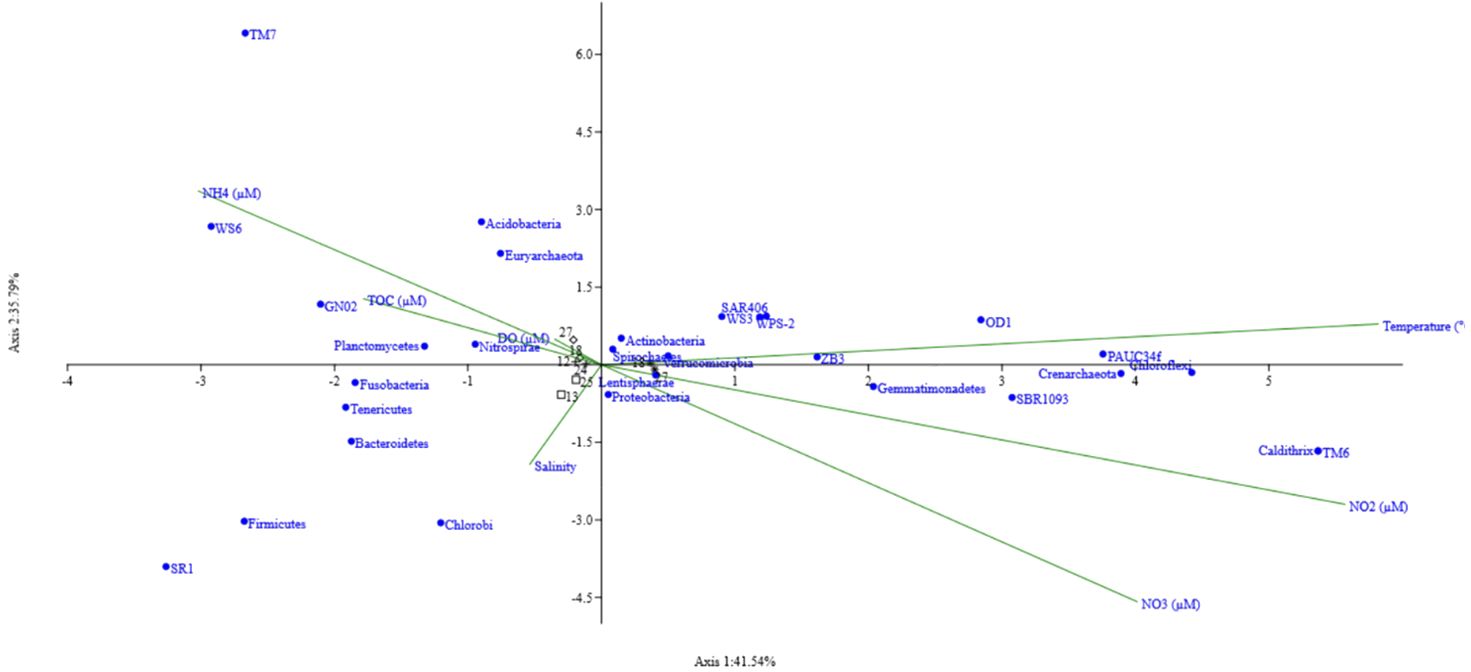
Figure 3 Canonical correspondence analysis ordination Triplot of bacterial communities at phyla level at the Candolim time-series station G5 associated with environmental variables. At the end of the spokes are the environmental parameters, black are the depths sampled in each cruise/field trip, and blue dots represent the bacterial phyla.
3.4 Predictive functional analysis
The functions predicted by Tax4fun2 highlighted various diverse functions performed by the organisms that are essential for numerous ecological processes. As mentioned, these inferences are predictive in nature, so need to be substantiated using more direct methods like metatranscriptome, metaproteomic or shot-gun sequencing. The dominant pathways reported were involved in metabolism, such as carbohydrate, lipid, energy, nucleotide, amino acid metabolism, genetic information processing (Supplementary Figure 3), cellular processes, and environmental information processing. Few of the abundant genes predicted in all the samples were sulfite reductase (K00392), thiosulfate sulfurtransferase (K01011), Nitrogen regulatory protein Pii1 (K02589), thioredoxin 1 (K03671), DNA repair and replication protein RecF (K03629) etc. (Supplementary Figure 3).
4 Discussion
The Western Shelf of India is considered one of the most productive areas of the Arabian Sea due to nutrient enrichment caused during the Southwest monsoonal upwelling (Naqvi et al., 2000, 2006; Shetye et al., 1990). This area experiences considerable changes in DO concentration throughout the year. It remains well-oxygenated from November-May, subsequently reaching hypoxic conditions by July and intensifying further during September-October. Hypoxia begins in June, followed by a decrease in the DO levels and an increase in denitrification during late August-early September, further progressing towards anoxia associated with sulphidic conditions during September-October (Naqvi et al., 2006, 2010).
The reduction of DO levels at the sampling site is a characteristic feature observed during the SWM (Naqvi et al., 2006) and is pretty evident in the study. Even in the present study, the conditions switched from hypoxic to suboxic to anoxic from July to October 2018. The reduction in the DO values may be attributed to the consumption of DO during the remineralization of the organic matter produced as a result of upwelling (Naqvi et al., 2000). Concomitant with the reduction in the DO levels, nitrogen is also lost during September – October due to denitrification. This is evident from the loss of nitrate and nitrite during these months. These results correspond to the studies conducted by Naqvi et al. (Naqvi et al., 2006) at the exact location. Unlike the open ocean Arabian Sea OMZ, which experiences only nitrogen loss due to denitrification and anammox (Ward et al., 2008, 2009; Lam et al., 2000), the Western Shelf of India also undergoes sulphate reduction (complete anoxia) as seen from the presence of H2S (1.66 µM) in the near-bottom depths during October 2018, which was also reported previously (Naqvi et al., 2006; Shenoy et al., 2012; Pratihary et al., 2014). One of the outcomes of such a condition is the accumulation of unprecedented amounts of nitrous oxide (~ 1 µM, a greenhouse gas (Naqvi et al., 2006), and dimethylsulphide (~ 0.5 µM, an anti-greenhouse gas) (Shenoy et al., 2012). However, the sulfidic conditions have been reported to occur during September-October, followed by the release of nitrogen from the decaying organic matter and its accumulation as ammonium (Naqvi et al., 2000, 2006).
Microbial communities are complex; their structure and functions tend to change in response to various physicochemical and environmental factors. Most environmental factors affecting microbial communities and their structural dynamics differ according to time and space (Fuhrman et al., 2015). The effect of these variables on the diversity of prokaryotic communities is reported in the present study. Prokaryotic communities in the study area have been known to exhibit a distinct seasonal pattern. Several environmental variables significantly influence the differences in abundance and distribution patterns. The bacterial communities observed in the study area were similar to those reported from coastal and open ocean oxygen-deficient regions (Wright et al., 2012; Fernandes et al., 2019, 2020). Proteobacteria dominated the bacterial community structure at the phylum level in most samples, similar to the results obtained previously by Gomes et al. (2019). The other phyla observed included Actinobacteria, Bacteroidetes, Chloroflexi, and Planctomycetes, similar to those identified previously in oxygen minimum zones (Fuhrman et al., 2015; Gomes et al., 2019). Bacteroidetes contribute significantly to the marine bacterial community and are dominant in nutrient-rich waters (Kirchman, 2002). Chloroflexi and Planctomycetes reported in the study area have been previously reported in the hypoxic regions and are known for breaking down sulfated heteropolysaccharides, denitrification and carbon fixation, respectively (Ye et al., 2016; Yilmaz et al., 2016). Verrucomicrobia, a minor group present in all the samples at both depths, is known to be ubiquitous in marine environments, and its higher relative abundance in July 2018 and October 2019 seems to indicate their affinity for oxic waters.
At the class level, Acidimicrobia belonging to phyla Actinobacteriota was dominant in both the depths in all the samples. The classes Alphaproteobacteria and Gammaproteobacteria were also ubiquitously present in all the samples. These Proteobacteria classes have been known to account for 30 - 50% of all bacteria in the surface waters in the Oceans (Gomes et al., 2019). The presence of Firmicutes reported in the study coincides with the findings of Gomes et al. (2020), and these are known to encompass efficient denitrifiers. The predominant class, Acidobacteria, is known to be involved in nitrate reduction (Kielak et al., 2016).
The archaeal community was dominated by Euryarchaeal class Thermoplasmata, marine group II and marine group III in all the samples. A higher abundance of marine group II Euryarchaea observed in these hypoxic and anoxic waters was parallel to that reported in Arabian Sea OMZ, in the black sea, and anoxic Cariaco Basin, wherein this phylum was present throughout the water column (Madrid et al., 2001; Vetriani et al., 2003; Bandekar et al., 2018). Marine group II archaea have been known to play an important role in carbon cycling (Zhang et al., 2015). The higher relative abundance of Euryarchaea during October 2018 indicates its affinity to thrive in anoxic waters, as reported previously by Belmar et al. (2011). It has also been suggested that the concurrent aerobic and anaerobic processes in oxygen-deficient zones may lead to higher archaeal diversity in those regions (Füssel, 2013). Nanoarchaeato was reported from all the water samples, though at very low abundance.
The various genera in all the samples have been known to play a significant role in biogeochemical cycles. The OM60/nor five clades belonging to the class Gammaproteobacteria are aerobic anoxygenic photoheterotrophs that play an important role in the carbon cycle (Yan, 2009). Similarly, the SAR324 clade of class Deltaproteobacteria are potential sulfur oxidizers and also participate in carbon fixation (Sheik et al., 2014). The presence of sulfur oxidizing marine chemoautotrophs SUP05 clade in almost all is concurrent with the findings of Canfield et al. (2010). This group of bacteria are known to dominate suboxic and anoxic waters and is assumed to take part in denitrification and annamox processes (Walsh et al., 2009; Hawley et al., 2014).
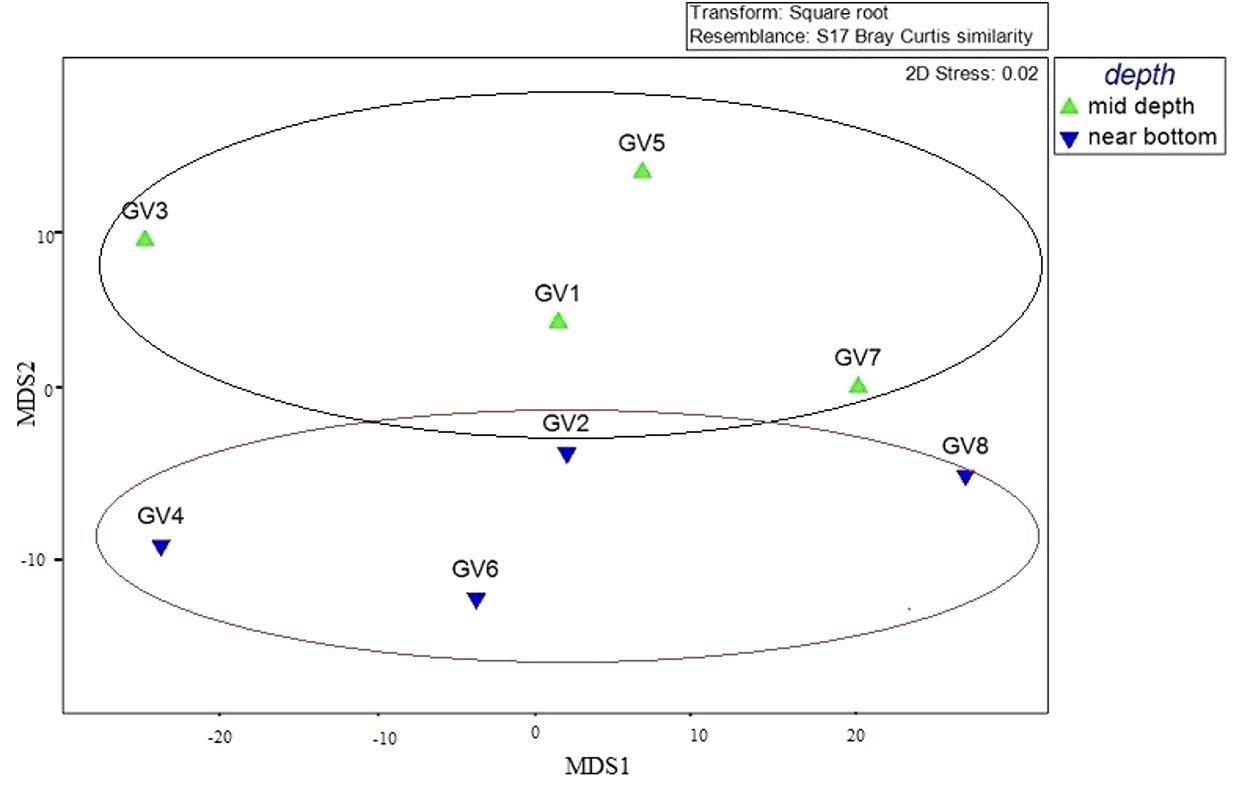
MDS and PCoA analyses based on Bray-Curtis distance at phyla level based on depth showed that the hierarchical clustering formed groups of mid and near-bottom depth samples (Figure 4) and (Supplementary Figure 1). In addition, the MDS analysis showed clustering of samples based on depths and DO levels. GV1, GV2, GV7 and GV8 are closer to each other and represent higher oxygen content than GV3, GV4, GV5 and GV6. A similar partitioning pattern was observed between the surface, oxygen minimum zone and near-bottom depths in other known OMZs (Stevens and Ulloa, 2008). Among the five environmental variables used for correlation analysis, it was observed that DO, ammonia, nitrate and nitrite significantly affected the abundance of bacteria and the archaeal community. DO is considered crucial in determining changes in the microbial community composition in the marine environment (Stevens and Ulloa, 2008), which was also evident in this study, with the community structure changing with the change in DO level. The functional analysis predicted using Tax4fun2 gave insights into the functional capabilities of microbial communities in the study area. This, in turn, highlighted the dominant pathways involved in various ecological processes, thus emphasizing the significant role played by the bacterial communities in the coastal hypoxic conditions.
One of the largest naturally occurring oxygen-deficient zone develops during the late southwest monsoon over the west coast of India, resulting from upwelling, which leads to the depletion of DO in that area (Naqvi et al., 2000). These low oxygen conditions are known to have severe effects on the coastal marine environment resulting in a loss of marine biodiversity. The prokaryotic communities are intensely affected by seasonal changes and respond by shifting community composition based on these environmental factors. The abundance and distribution of various microbial lineages can be studied using high-throughput sequencing methods. These studies will serve as a valuable tool to interpret the importance of different bacterial and archaeal groups in regulating biogeochemical cycling in hypoxic environments.
5 Conclusion
Coastal ecosystems are greatly influenced by anthropogenic activities, which can be attributed to growing industrial development and pollution. These activities have the potential to influence the natural flow of events, such as the development of seasonal hypoxia along the west coast of India. This may eventually add to the declining oxygen concentration in the coastal environments and change the ecological functioning. High throughput sequencing techniques can be used to study the changes in microbial diversity, which provides insights into the distribution of various microbial communities. Microorganisms contribute significantly to the biogeochemical cycling of several essential elements such as nitrogen, carbon, phosphate, and sulfur. Therefore, the influence of oxygen depletion on microbial community structure affects coastal environmental management and the global environment. Studying the implications of low oxygen concentration and other environmental variables on bacterial community structure provides insights into the processes that might be affected due to the development of hypoxia.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA 706932.
Author contributions
VN: Data curation, Formal analysis, Methodology, Validation, Writing – original draft, Software, Visualization. SD: Data curation, Formal analysis, Methodology, Validation, Writing – original draft, Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing. SS: Data curation, Formal analysis, Methodology, Software, Visualization, Writing – review & editing. DS: Funding acquisition, Supervision, Validation, Writing – review & editing. AM: Methodology, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The authors thank the Council of Scientific and Industrial Research (CSIR), Govt. of India for funding under the projects MLP1802 and OLP1901, and the Ministry of Earth Sciences (MoES) for funding the Time Series studies under project GAP2425. Shruti Shah thanks Council of Scientific and Research (CSIR) for her Fellowship (CSIR-SRF 31/26(0330)/2019-EMR-I).
Acknowledgments
The authors thank the Director of CSIR-National Institute of Oceanography for the support and infrastructure facilities. We also thank the reviewers for their suggestions and constructive comments, which substantially improved the manuscript. The authors thank the crew and the staff involved in the cruises SSD052 and SSD57 and the field trips CaTS 157 and CaTS 161.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1301955/full#supplementary-material
References
Bandekar M., Ramaiah N., Jain A., Meena R. M. (2018). Seasonal and depth-wise variations in bacterial and archaeal groups in the Arabian Sea oxygen minimum zone. Deep-Res Part II 156, 4–18. doi: 10.1016/j.dsr2.2017.12.015
Belmar L., Molina V., Ulloa O. (2011). Abundance and phylogenetic identity of archaeoplankton in the permanent oxygen minimum zone of the eastern tropical South Pacific. FEMS Microbiol. Ecol. 78, 314–326. doi: 10.1111/j.1574-6941.2011.01159.x
Beman J. M., Carolan M. T. (2013). Deoxygenation alters bacterial diversity and community composition in the ocean’s largest oxygen minimum zone. Nat. Commun. 4, 1–11. doi: 10.1038/ncomms3705
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Breitburg D., Levin L. A., Oschlies A., Grégoire M., Chavez F. P., Conley D. J. (2018). Declining oxygen in the global ocean and coastal waters. Science 359. doi: 10.1126/science.aam7240
Canfield D. E., Stewart F. J., Thamdrup B. O., Brabandere L. D., Dalsgaard T., Delong E. F., et al. (2010). A Cryptic Sulfur cycle in Oxygen-Minimum-zone waters off the Chilean coast. Science 330, 1375–1378. doi: 10.1126/science.1196889
Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Carpenter J. H. (1965). The Chesapeake Bay Institute technique for the Winkler dissolved oxygen method. Limnol Oceanogr 10, 1413. doi: 10.4319/lo.1965.10.1.0141
Clarke K. R., Somerfield P. J., Gorley R. N. (2008). Testing of null hypotheses in exploratory community analyses: similarity profiles and biota-environment linkage. J. Exp. Mar. Biol. Ecol. 366, 56–69. doi: 10.1016/j.jembe.2008.07.009
Diaz R. J., Rosenberg R. (2008). Spreading dead zones and consequences for marine ecosystems. Science 321, 926–929. doi: 10.1126/science.1156401
Fernandes G. L., Shenoy B. D., Damare S. R. (2020). Diversity of bacterial community in the oxygen minimum zones of Arabian Sea and Bay of Bengal as deduced by Illumina sequencing. Front. Microbiol. 153. doi: 10.3389/fmicb.2019.03153
Fernandes G. L., Shenoy B. D., Menezes L. D., Meena R. M., Damare S. R. (2019). Prokaryotic diversity in oxygen depleted waters of the Bay of Bengal inferred using culture-dependent and-independent methods. Ind. J. Microbiol. 59, 193–199. doi: 10.1007/s12088-019-00786-1
Fuhrman J. A., Cram J. A., Needham D. M. (2015). Marine microbial community dynamics and their ecological interpretation. Nat. Rev. Microbiol. 13, 133–146. doi: 10.1038/nrmicro3417
Füssel J. (2013). Impacts and importance of ammonia-and nitrite oxidation in the marine nitrogen cycle (Bremen: Max Planck Institute for Marine Microbiology).
Gomes J., Khandeparker R., Meena R. M., Ramaiah N. (2019). Bacterial community composition markedly altered by coastal hypoxia. Ind. J. Microbiol. 59, 200–208. doi: 10.1007/s12088-019-00790-5
Gomes J., Khandeparker R., Naik H., Shenoy D., Meena R. M., Ramaiah N. (2020). Denitrification rates of culturable bacteria from a coastal location turning temporally hypoxic. J. Mar. Syst. 209, 103089. doi: 10.1016/j.jmarsys.2018.06.003
Hammer Ø., Harper D. A., Ryan P. D. (2021). PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron 4, 9. Available at: https://palaeoelectronica.org/2001_1/past/issue1_01.htm.
Hawley A. K., Brewer H. M., Norbeck A. D., Pasa-Tolic L., Hallam S. J. (2014). Metaproteomics reveals differential modes of metabolic coupling among ubiquitous oxygen minimum zone microbes. PNAS 111, 11395–11400. doi: 10.1073/pnas.1322132111
Kielak A. M., Barreto C. C., Kowalchuk G. A., van Veen J. A., Kuramae E. E. (2016). The ecology of Acidobacteria: moving beyond genes and genomes. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00744
Kirchman D. L. (2002). The ecology of Cytophaga–Flavobacteria in aquatic environments. FEMS Microbial Ecol. 39, 91–100. doi: 10.1016/S0168-6496(01)00206-9
Lam P., Jensen M., Lavik G., McGinnis D., Muller B., Schubert C., et al. (2000). Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc. Natl. Acad. Sci. 104, 7104–7109. doi: 10.1073/pnas.0611081104
Lipsewers Y. A., Vasquez-Cardenas D., Seitaj D., Schauer R., Hidalgo-Martinez S., Damsté J. S. S., et al. (2017). Impact of seasonal hypoxia on activity and community structure of chemolithoautotrophic bacteria in a coastal sediment. Appl. Environ. Microbiol. 83, e03517-. doi: 10.1128/AEM.03517-16
Lynch M. D., Neufeld J. D. (2015). Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 13, 217–229. doi: 10.1038/nrmicro3400
Madhupratap M., Kumar S. P., Bhattathiri P. M. A., Kumar M. D., Raghukumar S., Nair K K. C., et al. (1996). Mechanism of the biological response to winter cooling in the north-eastern Arabian Sea. Nature 384, 549–552. doi: 10.1038/384549a0
Madrid V. M., Taylor G. T., Scranton M. I., Chistoserdov A. Y. (2001). Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67, 1663–1674. doi: 10.1128/AEM.67.4.1663-1674.2001
Magoč T., Salzberg S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
McDonald D., Price M. N., Goodrich J., Nawrocki E. P., DeSantis T. Z., Probst A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Naqvi S. W. A., Bange H. W., Farías L., Monteiro P. M. S., Scranton M. I., Zhang J. (2010). Marine hypoxia/anoxia as a source of CH4 and N2O. Biogeosciences 7, 2159–2190. doi: 10.5194/bg-7-2159-2010
Naqvi S. W. A., Jayakumar D. A., Narvekar P. V., Naik H., Sarma V. V. S. S., D’souza W., et al. (2000). Increased marine production of N2O due to intensifying anoxia on the Indian continental shelf. Nature 408, 346–349. doi: 10.1038/35042551
Naqvi S. W. A., Naik H., Pratihary A., Narvekar P. V., Jayakumar D. A., Devol A. H., et al. (2006). Coastal versus open-ocean denitrification in the Arabian Sea. Biogeosciences 3, 621–633. doi: 10.5194/bg-3-621-2006
Parks D. H., Tyson G. W., Hugenholtz P., Beiko R. G. (2014). STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Pratihary A. K., Naqvi S. W. A., Narvenkar G., Kurian S., Naik H., Naik R. (2014). Benthic mineralisation and nutrient exchange over the inner continental shelf of western India. Biogeosciences 11, 2771–2791. doi: 10.5194/bg-11-2771-2014
Rabalais N. N., Turner R. E., Wiseman W. J. Jr. (2002). Gulf of Mexico hypoxia, aka “The dead zone”. Annu. Rev. Ecol. Sys 33, 235–263. doi: 10.1146/annurev.ecolsys.33.010802.150513
Reed H. E., Martiny J. B. (2013). Microbial composition affects the functioning of estuarine sediments. ISME J. 7, 868–879. doi: 10.1038/ismej.2012.154
Sheik C. S., Jain S., Dick G. J. (2014). Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ. Microbiol. 16, 304–317. doi: 10.1111/1462-2920.12165
Shenoy D. M., Sujith K. B., Gauns M. U., Patil S., Sarkar A., Naik H. (2012). Production of dimethylsulphide during the seasonal anoxia off Goa. Biogeochemistry 110, 47–55. doi: 10.1007/s10533-012-9720-5
Shetye S. R., Gouveia A. D., Shenoi S. S. C., Sundar D., Michael G. S., Almeida A. M., et al. (1990). Hydrography and circulation off the west coast of India during the southwest monsoon 1987. J. Mar. Res. 48, 359–378. doi: 10.1357/002224090784988809
Spietz R. L., Williams C. M., Rocap G., Horner-Devine M. C. (2015). A dissolved oxygen threshold for shifts in bacterial community structure in a seasonally hypoxic estuary. PloS One 10, e0135731. doi: 10.1371/journal.pone.0135731
Stevens H., Ulloa O. (2008). Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ. Microbiol. 10, 1244–1259. doi: 10.1111/j.1462-2920.2007.01539.x
Sudheesh V., Gupta G. V. M., Naqvi S. W. A. (2020). Massive methane loss during seasonal hypoxia/anoxia in the nearshore waters of south-eastern Arabian Sea. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00324
Thamdrup B. O., Rosselló-Mora R., Amann R. (2000). Microbial manganese and sulfate reduction in Black Sea shelf sediments. Appl. Environ. Microbiol. 66, 2888–2897. doi: 10.1128/AEM.66.7.2888-2897.2000
Vetriani C., Tran H. V., Kerkhof L. J. (2003). Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl. Environ. Microbiol. 69, 6481–6488. doi: 10.1128/AEM.69.11.6481-6488.2003
Walsh D. A., Zaikova E., Howes C. G., Song C. Y., Wright J. J., Tringe S. G., et al. (2009). Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326, 578–582. doi: 10.1126/science.1175309
Ward B., Devol A., Rich J., Chang B., Bulow S., Naik H., et al. (2009). Denitrification as the dominant nitrogen loss process in the Arabian Sea. Nature 461, 78–81. doi: 10.1038/nature08276
Ward B. B., Tuit C. B., Jayakumar A., Rich J. J., Moffett J., Naqvi S. W. (2008). Organic carbon, and not copper, controls denitrification in oxygen minimum zones of the ocean. Deep-Sea Res. Part I 55, 1672–1683. doi: 10.1016/j.dsr.2008.07.005
Wemheuer F., Taylor J. A., Daniel R., Johnston E., Meinicke P., Thomas T., et al. (2020). Tax4Fun2 predicts habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 15, 11. doi: 10.1186/s40793-020-00358-7
Wiggert J. D., Hood R. R., Banse K., Kindle J. C. (2005). Monsoon-driven biogeochemical processes in the Arabian Sea. Prog. Oceanogr 65, 176–213. doi: 10.1016/j.pocean.2005.03.008
Wright J. J., Konwar K. M., Hallam S. J. (2012). Microbial ecology of expanding oxygen minimum zones. Nat. Rev. Microbiol. 10, 381–394. doi: 10.1038/nrmicro2778
Yan S. (2009). Molecular ecology of the nor5/OM60 group of gammaproteobacteria (Bremen: University of Bremen).
Ye Q., Wu Y., Zhu Z., Wang X., Li Z., Zhang J. (2016). Bacterial diversity in the surface sediments of the hypoxic zone near the Changjiang Estuary and in the East China Sea. Microbiol. Open 5, 323–339. doi: 10.1002/mbo3.330
Yilmaz P., Yarza P., Rapp J. Z., Glöckner F. O. (2016). Expanding the world of marine bacterial and archaeal clades. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01524
Keywords: 16S rRNA, amplicon sequence variants (ASVs), Candolim Time-series Station (CaTS), hypoxia, Illumina MiSeq, southwest monsoon
Citation: Naik VV, Damare SR, Shah SS, Shenoy DM and Mulla AB (2024) Effect of coastal hypoxia on bacterial diversity as elucidated through 16S rRNA amplicon sequencing. Front. Mar. Sci. 11:1301955. doi: 10.3389/fmars.2024.1301955
Received: 25 September 2023; Accepted: 24 April 2024;
Published: 13 May 2024.
Edited by:
Anne Bernhard, Connecticut College, United StatesReviewed by:
Stefan Andrei, Academy of Sciences of the Czech Republic (ASCR), CzechiaLina Blandon, National University of Colombia, Colombia
Copyright © 2024 Naik, Damare, Shah, Shenoy and Mulla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Samir R. Damare, samir@nio.org
†Present address: Amara Begum Mulla, Department of Biotechnology, Dhempe College of Arts and Science, Miramar, Goa, India
‡ORCID: Samir R. Damare, orcid.org/0000-0002-4201-4844
Shruti S. Shah, orcid.org/0000-0002-0515-5648
 Vruti V. Naik1
Vruti V. Naik1  Samir R. Damare
Samir R. Damare Shruti S. Shah
Shruti S. Shah Damodar M. Shenoy
Damodar M. Shenoy